90 90.90
RESEARCH ARTICLE SUMMARY ◥
lease of the TetR-Clr4 initiator (TetR-Clr4-I)from DNA.
RESULTS: Cells containing the reporter
gene in combination with the expressionof TetR-Clr4-I formed pink colonies on low-
Epigenetic inheritance uncoupled
adenine medium lacking tetracycline, in-dicating ade6+ silencing. The establishment
from sequence-specific recruitment
of heterochromatin resulted in high levelsof H3K9 methylation (H3K9me), which was
subsequently lost upon
Kaushik Ragunathan, Gloria Jih, Danesh Moazed*
tetracycline-induced re-lease of TetR-Clr4-I within
Read the full article
INTRODUCTION: Changes in histone post-
RATIONALE: The fission yeast Schizosaccharo-
at http://dx.doi.
10 cell divisions, result-
translational modifications are associated
myces pombe contains chromosomal domains
ing in the appearance of
with epigenetic states that define distinct pat-
that share many features with heterochroma-
white colonies. Whereas
terns of gene expression. Whereas sequence-
tin in multicellular eukaryotes, such as meth-
perturbations to path-
specific DNA binding proteins play essential
ylation of histone H3 lysine 9 (H3K9), catalysis
ways that altered the rate of histone exchange
roles in establishing an epigenetic state, their
by the human Suv39h homolog Clr4, asso-
or eliminating competition from endoge-
contributions to maintenance remain unclear.
ciation with HP1 proteins (Swi6 and Chp2),
nous heterochromatic loci had subtle effects
Previous attempts to separate the inheritance
and histone hypoacetylation. We developed
on epigenetic inheritance of ade6+ silenc-
of epigenetic states from sequence-specific
an inducible system for heterochromatin es-
ing, deletion of the putative JmjC domain–
establishment suggest that specific DNA se-
tablishment in S. pombe by fusion of the Clr4
containing demethylase Epe1 resulted in
quences and DNA binding proteins are con-
methyltransferase catalytic domain to the bac-
cells that retained ade6+ silencing for >50
tinuously required for epigenetic inheritance.
terial tetracycline repressor (TetR) protein. To
generations after tetracycline-induced re-
Moreover, in addition to DNA binding pro-
generate a reporter locus, we introduced 10
lease of TetR-Clr4-I or deletion of the TetR
teins, the establishment and maintenance of
tetracycline operators upstream of the normal-
module. Furthermore, the chromodomain of
epigenetic states involves self-reinforcing in-
ly expressed ade6+ gene (10XtetO-ade6+). The
Clr4, which is involved in recognition of the
teractions between histone modifications and
silencing of ade6+ results in the formation
H3K9me mark, was indispensable for main-
RNA interference (RNAi) or DNA methylation.
of red or pink colonies upon growth on me-
tenance, suggesting that a direct "read-write"
Therefore, whether histone-based mechanisms
dium with limiting adenine concentrations.
mechanism mediated by Clr4 propagates
can transmit epigenetic memory indepen-
This system allowed us to determine whether
histone modifications and allows histones
dently of specific DNA sequences remains
heterochromatin, once established, could be
to act as carriers of epigenetic information.
maintained after tetracycline-mediated re-
This mechanism allows epigenetic statesto be inherited during mitosis and meiosisand is also critical for maintaining low levels
of H3K9me at native pericentromeric repeats.
CONCLUSION: Our findings indicate that
even in the absence of any coupling to otherpositive-feedback loops, or in the absence
of sequence-dependent initiation signals,H3K9me defines a silent state that can be
epigenetically inherited. Maintenance ofthe OFF state is determined by the balance
between the rate of H3K9me by the Clr4reader-writer module and the loss rate due to
clr4+, epe1+
demethylation by an Epe1-dependent mech-anism, transcription-coupled nucleosome ex-
clr4+, epe1∆
change, and dilution of histones during DNAreplication. The regulation of histone de-
red colony = silent, white colony = expressed
methylation activity may play a broad role indetermining the reversibility of epigenetic
states.
▪
RELATED ITEMS IN SCIENCE
P. N. C. B. Audergon et al., Restrictedepigenetic inheritance of H3K9 methylation.
Science 348, 132
–135 (2015).
H3K9me defines a silent state that can be epigenetically inherited. A direct read-write
Department of Cell Biology, Howard Hughes Medical Institute,Harvard Medical School, 240 Longwood Avenue, Boston, MA
mechanism involving the Clr4 H3K9 methyltransferase propagates histone modifications and allows
histones to act as carriers of epigenetic information in the absence of any input from the DNA
*Corresponding author:
[email protected]
sequence, DNA methylation, or RNAi. Epe1, a putative demethylase, and other transcription-
Cite this article as K. Ragunathan et al., Science 348, 1258699(2015). DOI: 10.1126/science.1258699
associated histone turnover pathways modulate the rate of decay of the epigenetic state.
3 APRIL 2015 • VOL 348 ISSUE 6230
sciencemag.org SCIENCE
RESEARCH ARTICLE
◥
that domains of H3K9me can be inherited for >50generations in the absence of sequence-specificrecruitment and define central roles for the pu-tative demethylase, Epe1, in the erasure of H3K9me
and the chromodomain of the Clr4 methyltrans-ferase in its maintenance.
Epigenetic inheritance uncoupled
Inducible establishmentof heterochromatin
from sequence-specific recruitment
Silent chromatin domains can be established byectopic recruitment of histone-modifying enzymesto chromatin via fusion with heterologous DNA
Kaushik Ragunathan, Gloria Jih, Danesh Moazed*
binding proteins (, ). To create an induciblesystem for heterochromatin formation, we fused
Changes in histone posttranslational modifications are associated with epigenetic states
a Clr4 protein lacking its N-terminal chromodo-
that define distinct patterns of gene expression. It remains unclear whether epigenetic
main (required for binding to methylated histone
information can be transmitted through histone modifications independently of specific
H3K9) while retaining its enzymatic methyltrans-
DNA sequence, DNA methylation, or RNA interference. Here we show that, in the fission
ferase activity to the bacterial TetR protein (de-
yeast Schizosaccharomyces pombe, ectopically induced domains of histone H3 lysine
signated "TetR-Clr4-I" for TetR-Clr4 initiator)
9 methylation (H3K9me), a conserved marker of heterochromatin, are inherited through
(Fig. 1A). The TetR DNA binding domain facil-
several mitotic and meiotic cell divisions after removal of the sequence-specific initiator.
itates protein targeting to a locus that harbors
The putative JmjC domain H3K9 demethylase, Epe1, and the chromodomain of the H3K9
its cognate DNA binding sequence, and this re-
methyltransferase, Clr4/Suv39h, play opposing roles in maintaining silent H3K9me
cruitment activity is abrogated by the addition
domains. These results demonstrate how a direct "read-write" mechanism involving
of tetracycline (TetRoff system) We gener-
Clr4 propagates histone modifications and allows histones to act as carriers of
ated cells in which TetR-Clr4-I replaced the wild-
type (WT) Clr4 (TetR-clr4-I) or in which WT Clr4
was intact and TetR-Clr4-I was inserted at an-
n individual cell can give rise to progeny
response element, which acts analogously to
other locus (TetR-clr4-I, clr4+). Comparisons be-
with distinct patterns of gene expression
the yeast silencer, is continuously required for
tween strains with or without clr4+ allowed us
and phenotypes without change in its DNA
maintenance of domains of histone H3 lysine 27
to evaluate the contribution of the Clr4 chromo-
sequence. In eukaryotic cells, a major mech-
(H3K27) methylation and reporter gene silencing
domain to establishment and/or maintenance.
anism that gives rise to such phenotypic
, ). The question therefore remains as to
To generate a reporter locus, we replaced the eu-
or epigenetic states involves changes in histone
whether histones can act as carriers of epigenetic
chromatic ura4+ locus with an ade6+ gene con-
posttranslational modifications and chromatin
information in the absence of any input from the
taining 10 tetracycline operators immediately
structure , ). The basic unit of chromatin is the
underlying DNA sequence.
upstream of the promoter (10XtetO-ade6+) (Fig.
nucleosome, which is composed of 147 base pairs
The fission yeast Schizosaccharomyces pombe
1A). ade6+ provides a convenient visual reporter,
(bp) of DNA wrapped twice around an octamer
contains extensive domains of heterochromatin
as its silencing results in formation of red or pink
composed of histones H2A, H2B, H3, and H4 (
at its pericentromeric DNA repeats, subtelomeric
colonies upon growth on medium with limiting
The highly conserved basic N termini and, to a
regions, and the silent mating type loci (These
adenine concentrations (
lesser extent, the globular domains of histones
domains share many features of heterochromatin
As shown in Fig. 1B, cells containing the
contain a variety of posttranslational modifica-
in multicellular eukaryotes, such as H3K9 meth-
10XtetO-ade6+ reporter in combination with the
tions that affect nucleosome stability or provide
ylation (H3K9me), which is catalyzed by the hu-
expression of the TetR-Clr4-I fusion protein, but
binding sites for effectors that activate or repress
man Suv39h homolog Clr4; association with HP1
not those containing the fusion protein alone or
proteins (Swi6 and Chp2); and histone hypoacet-
the reporter alone, formed pink colonies on low-
It has been established for nearly four decades
ylation. Furthermore, S. pombe heterochromatin
adenine medium lacking tetracycline (–tet). Con-
that parental histones are retained and randomly
displays epigenetic inheritance properties in which
sistent with previous observations (silencing
distributed to newly synthesized daughter DNA
cells containing a reporter gene inserted within
did not require clr4+—suggesting that the chromo-
strands during DNA replication (It was
heterochromatin display variegating reporter gene
domain of Clr4 was not required for de novo
therefore logical to propose that combinations
expression The ON and OFF states of such
heterochromatin establishment (Fig. 1B)—but
of histone modifications, sometimes referred to
reporter genes can be stably transmitted in cis
depended on HP1 proteins (Swi6 and Chp2) and
as a "histone code," are responsible for epigenetic
through both mitotic and meiotic cell divisions
histone deacetylases (Clr3 and Sir2), which act
memory of gene expression patterns , How-
(However, because these observations of
downstream of H3K9me (fig. S1A). Colonies grown
ever, previous attempts to separate sequence-
epigenetic inheritance were made at native se-
in the absence of tetracycline were then plated
specific establishment from maintenance have
quences, contributions arising from sequence-
on medium containing tetracycline (+tet) to de-
failed to provide unambiguous support for a pure-
specific elements that stabilize heterochromatin
termine whether the silent state could be main-
ly histone-based inheritance mechanism in sys-
could not be ruled out To determine whether
tained upon release of TetR-Clr4-I from DNA.
tems that display stable epigenetic expression
heterochromatin maintenance can be separated
Tetracycline-dependent release of TetR-Clr4-I from
states , ). Most notably, silencers (i.e., DNA
from the sequences that initiate its establishment,
tetO sites resulted in the loss of silencing, as in-
sequences that mediate the establishment of
we developed a system for inducible heterochro-
dicated by the formation of white colonies (Fig.
hypoacetylated domains of silent chromatin in
matin establishment in S. pombe by fusion of
1B, +tet). Chromatin immunoprecipitation (ChIP)
the budding yeast Saccharomyces cerevisiae) are
the Clr4 methyltransferase catalytic domain to
experiments verified that tetracycline addition
continuously required for maintenance of the si-
the bacterial tetracycline repressor (TetR) pro-
resulted in release of TetR-Clr4-I from the 10XtetO
lent state ). Similarly, the Drosophila Polycomb
tein. This allowed us to establish an extended
sites, as the TetR ChIP signal in the presence of
heterochromatic domain and study its initiator-
tetracycline was near background levels sim-
Department of Cell Biology, Howard Hughes Medical
independent maintenance either by tetracycline-
ilar to that observed for cells lacking TetR-Clr4-I
Institute, Harvard Medical School, 240 Longwood Avenue,
mediated release of TetR-Clr4 from DNA or after
(Fig. 1C). Furthermore, ChIP combined with high-
Boston, MA 02115, USA.
*Corresponding author:
[email protected]
deletion of the TetR module. Our results indicate
throughput sequencing (ChIP-seq) and ChIP
SCIENCE sciencemag.org
3 APRIL 2015 • VOL 348 ISSUE 6230
RESEARCH RESEARCH ARTICLE
quantitative polymerase chain reaction (qPCR)
but not the Dicer ribonuclease (Dcr1) or the
resulted in the formation of only white colonies
experiments showed that a 40- to 50-kb domain
Argonaute protein (Ago1) (Fig. 3, B and C). Thus,
on +tet medium (Fig. 3E). Therefore, rather than
of H3K9 di- and trimethylation (H3K9me2 and
maintenance requires the machinery that acts
differences in clr4+ dosage, the appearance of red
-me3) encompassing the 10XtetO-ade6+ region
downstream of H3K9me but occurs indepen-
or sectored colonies depends on the presence of
was completely lost 24 hours ( 10 cell divisions)
dently of an RNA interference (RNAi)–based
Clr4 with an intact chromodomain.
after the addition of tetracycline to the growth
mechanism. Consistent with the idea that Epe1
In an attempt to identify other features of
medium (Fig. 1D and fig. S1, B to D). We obtained
acts as a demethylase, the replacement of epe1+
chromatin that could be important for mainte-
similar results with cells that carried a WT copy
with alleles containing active-site mutations (epe1-
nance, we deleted two genes that are associated
of clr4+ in addition to TetR-clr4-I (Fig. 1D and
K314A and epe1-H297A) , ) displayed a main-
with transcription: mst2+, which encodes a his-
fig. S1D, lower two rows). These results demonstrate
tenance phenotype similar to its deletion (epe1D)
tone acetyltransferase (), and set1+, which en-
that a domain of H3K9me and the associated
(Fig. 3D). Furthermore, consistent with a require-
codes a histone H3K4 methyltransferase (In
silent state are reversed within 10 cell divisions
ment for the chromodomain of clr4+ in mainte-
both cases, we observed establishment that was
after release of the sequence-specific initiator
nance (Fig. 2A), the replacement of clr4+ with a
stronger than that of the wild type (–tet), as well
clr4-ID allele (which lacks the chromodomain)
as weak maintenance (+tet) effects (fig. S4, A and
Inheritance uncoupled fromsequence-specific initiation
A minimal mechanistic requirement for inheri-tance of a domain containing H3K9me marks
involves the recognition of preexisting H3K9
methylated histones coupled to the modifica-tion of newly deposited histones. The loss of a 45-kb domain of H3K9 dimethylation after
release of TetR-Clr4-I (Fig. 1D) may either be dueto nucleosome exchange processes associated
FLAG ChIP (24 hours)
with transcription and chromatin remodeling or
erasure of the methyl mark by a demethylase.
Alternatively, the rapid decay of methylation inthis domain may be facilitated by competition
between the ectopic locus and native heterochro-
ade6 TetR-clr4-I, clr4+
matic domains for a limiting pool of proteins that
play critical roles in maintenance.
To test these different scenarios, we constructed
TetR-clr4-I, 10XtetO-ade6+ cells (with or without
clr4+) that carried deletions for genes involvedin various chromatin-maintenance pathways
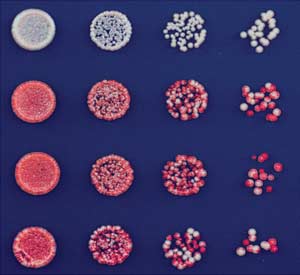
). The deletion of epe1+, which encodes a
domain size 45kb
putative histone H3K9 demethylase (), didnot affect the establishment of silencing, as in-
dicated by the appearance of red colonies on–tet medium for both TetR-clr4-I, epe1D cells and
TetR-clr4-I, clr4+, epe1D cells (Fig. 2A, left side).
In contrast to epe1+ cells, epe1D cells formed red,white, and sectored colonies in the presence of
TetR-clr4-I + tet
tetracycline, indicating that the silent state was
maintained after release of the initiator and thatthis phenotype was observed only in the case
TetR-clr4-I, clr4+ + tet
of cells that also contained a WT copy of clr4+(Fig. 2A, right side; compare last two rows withfirst two rows). ChIP experiments verified that
tetracycline addition promoted the release of
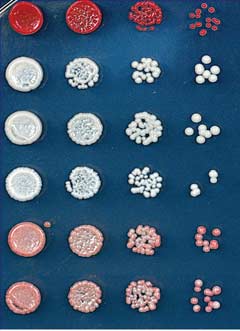
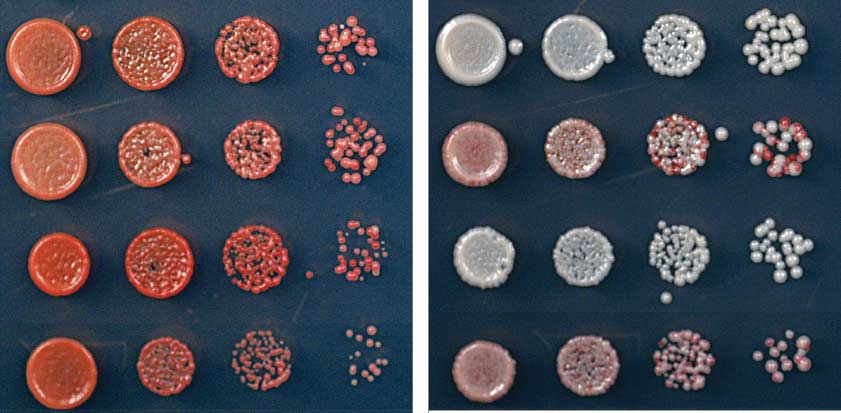
Fig. 1. Ectopic heterochromatin is lost after sequence-specific establishment upon tetracycline
TetR-Clr4-I from tetO sites (Fig. 2B). Consistent
addition. (A) Diagram of experimental scheme for TetR-Clr4-I (TetR-Clr4-I)–mediated H3K9me at the
with the colony color–silencing assays, ChIP-seq
ura4D::10XtetO-ade6+ locus. Tetracycline (tet) promotes the release of TetR-Clr4-I from tetO sites so that
and ChIP-qPCR experiments also indicated that
initiator-independent maintenance could be tested. (B) Test of ade6+ silencing on low-adenine medium
the large domain of H3K9 di- and trimethyla-
in the absence (-tet) and presence (+tet) of tetracycline. Silencing of ade6+ results in formation of red colonies.
tion surrounding the 10XtetO-ade6+ locus was lost
Centromeric ade6+ (otr1R::ade6) served as a positive control; the target locus alone (ura4D::10XtetO-
in TetR-clr4-I, epe1D cells but was maintained
ade6+) and ura4D::ade6+ cells served as negative controls. (C) ChIP-qPCR experiments assess the
in TetR-clr4-I, clr4+, epe1D cells (Fig. 2C and fig.
association of FLAG-tagged TetR-Clr4-I with the 10XtetO-ade6+ locus in the presence and absence of
S2, A to C) 24 hours after the addition of tetra-
tetracycline. The dash on the x axis indicates background ChIP signal from cells that did not express TetR-
cycline. Within this domain, the expression of
Clr4-I. In the presence of tetracycline, TetR-Clr4-I occupancy is close to background levels. Error bars
several other transcription units was silenced
indicate SD. (D) ChIP-seq experiments show that TetR-Clr4-I induces a de novo H3K9me2 domain that
on –tet medium, and this silencing was main-
surrounds the tetO-ade6+ locus (highlighted in red) for 20 kb on either side, in cells with or without clr4+,
tained for several hours after tetracycline addi-
which is lost 24 hours after tetracycline addition. H3K9me2 ChIP-qPCR data and H3K9me3 for
tion (fig. S3).
samples in (D) are presented in fig. S1. Chromosome 3 (site of insertion of 10XtetO-ade6+) and chro-
Maintenance of the silent state in epe1D cells
mosome 1 [centromere 1 left (cen1L)] coordinates are shown above the tracks, and read numbers (per
required the HP1 proteins (Swi6 and Chp2) and
million) are indicated on the right. H3K9me2 at DNA repeats of cen1L serves as an internal control for
the Clr3 and Sir2 histone deacetylases (Fig. 3A)
the ChIP-seq data.
3 APRIL 2015 • VOL 348 ISSUE 6230
sciencemag.org SCIENCE
RESEARCH RESEARCH ARTICLE
D). Although there were no obvious effects on
not correlate with the strength of the initial si-
for a subset of these time points are presented
colony color after 3 days of growth on tetracycline-
lenced state (establishment), indicating that per-
in Fig. 4, B to E, and the results for all time points
containing medium, we observed clear persist-
turbations to each pathway results in the release
are plotted in Fig. 4F. In general, upon tetra-
ence of H3K9me coupled with the release of
or recruitment of subsets of proteins that make dif-
cycline addition, the pattern of silencing of the
TerR-Clr4-I in mst2D cells 24 hours after the ad-
ferent contributions to the establishment and/or
10XtetO-ura4-GFP reporter in either epe1+ or epe1D
dition of tetracycline (fig. S4, B and C). We then
maintenance of heterochromatin.
over 2 days was consistent with the 10XtetO-
tested whether destabilizing endogenous het-
Because the different deletions essentially al-
ade6+ silencing results (Fig. 4, B and C). With the
erochromatic regions could release factors that
tered the rate of decay of ectopic H3K9me, we
higher time resolution and sensitivity of detect-
affect maintenance by deleting poz1+, a DNA bind-
hypothesized that epigenetic maintenance at the
ing phenotypic expression states, we found that
ing protein required for heterochromatin forma-
ectopic locus might also exist in WT cells over
the OFF state persists in 10% of the TetR-clr4-I,
tion at telomeres or dcr1+, which is required
the time scales of a few cell divisions. To observe
clr4+, epe1+ cells for up to 40 hours ( 20 cell divi-
for RNAi-dependent heterochromatin formation
the decay of silent chromatin with higher time
sions) after transfer to tetracycline-containing
at centromeres ((fig. S5). In these mutants,
resolution, we generated cells in which a green
medium (Fig. 4, D and F), whereas cells contain-
effects on colony color were absent (dcr1D, fig.
fluorescent protein (GFP) reporter was silenced
ing TetR-clr4-I, clr4+, epe1D displayed the most
S5A) or subtle (poz1D, fig. S5D), but we clearly
by TetR-Clr4-I. We modified the endogenous ura4+
stable maintenance patterns with 60% of the cells
observed persistence of H3K9me 24 hours after
gene to encode a Ura4-GFP fusion protein. In ad-
maintaining the OFF state, even after 100 hours
release of TetR-Clr4-I in both deletion backgrounds
dition, we inserted the same 10XtetO sites used
of growth ( 50 cell divisions) in tetracycline-
(fig. S5, B, C, E, and F).
in the earlier experiments immediately upstream
containing medium (Fig. 4E). These observa-
These results indicate that a domain of H3K9
of a 114-bp fragment of the ura4+ promoter (Fig.
tions indicate that epigenetic maintenance was
methylation and its associated silent state can
4A). As determined by fluorescence-activated cell
not unique to epe1D cells and that its detection
be maintained through mitotic cell divisions in
sorting (FACS) analysis, 10XtetO-ura4-GFP was
was normally masked by the rapid erasure of
the absence of sequence-specific initiation. The
silenced in a TetR-Clr4-I–dependent manner in
H3K9me marks by Epe1. Although the decay
decay rate of this epigenetic state is primarily in-
the presence or absence of clr4+. To assess decay
rate for cells containing TetR-clr4-I, epe1+ was
fluenced by the erasure of the methyl mark by
rates, we transferred cells that were grown in
rapid (<6 hours), the deletion of Epe1, even in
the putative demethylase Epe1 and, to a lesser ex-
medium lacking tetracycline (GFP OFF cells) to
this background, resulted in a slower decay rate
tent, by pathways that promote transcription or
medium containing tetracycline and harvested
(Fig. 4E). We observed the GFP OFF state in TetR-
heterochromatin assembly at endogenous loci.
samples at 0, 3, 6, 9, 12, 15, 23, 26, 32, 36, 39, 50,
clr4-I, epe1D cells lacking clr4+, 6 hours after trans-
We note that the efficiency of maintenance does
60, 77, and 100 hours for FACS analysis. FACS data
fer to tetracycline medium with a complete shiftto the ON state occurring only at 23 hours aftertransfer (Fig. 4C). In these cells, upon elimina-
FLAG ChIP (24 hours)
tion of Epe1, the decay rate is probably defined
primarily by the dilution of modified histones,
which may maintain epigenetic states for a fewgenerations in the absence of Clr4-mediated
It may be argued that maintenance of H3K9me
and the silent state in the initiator-based experi-ments might arise from low-affinity binding of
epe1 TetR-clr4-I, clr4+
TetR-Clr4-I to DNA in the presence of tetracy-
cline (We sought to unequivocally rule out
any role for sequence-dependent initiation in the
inheritance we observed by using homologousrecombination to replace TetR-clr4-I with clr4-
ID, which harbors a deletion of the TetR DNA
domain size 45kb
binding domain (Fig. 5A). After transformationto replace TetR-clr4-I with clr4-ID and plating on
selective low-adenine medium (Fig. 5A), we ob-
tained red, sectored, and white colonies, whichwere tested and confirmed for the replacement
event by allele-specific PCR (fig. S6). The isola-
TetR-clr4-I + tet
tion of red clr4-ID, epe1D cells confirmed that the
silent state could be maintained in the complete
absence of sequence-dependent Clr4 recruitment
TetR-clr4-I,clr4+ + tet
to DNA. Furthermore, the plating of red clr4-ID,epe1D cells on low-adenine medium produced red,sectored, and white colonies (Fig. 5B), whereas
plating of white clr4-I
D, epe1D isolates produced
only white colonies (Fig. 5C). In the absence of
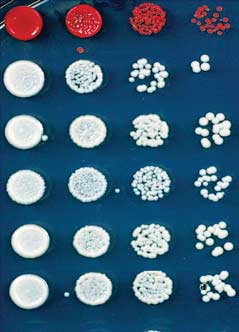
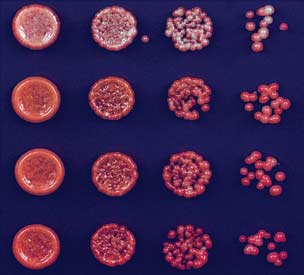
Fig. 2. Deletion of epe1+ allows maintenance of heterochromatin after release of TetR-Clr4-I.
the initiator and no other means of reestablish-
(A) Color-silencing assays showing that in TetR-clr4-I, clr4+, epe1D cells, silencing is maintained on
ment, the loss of the silent state was an irreversible
+tet medium. (B) ChIP experiments showing that TetR-Clr4-I is released from tetO sites in +tet medium.
event (Fig. 5C). Consistent with the deletion of
Error bars indicate SD. (C) ChIP-seq experiments showing that in epe1D cells, H3K9me2 is maintained
the TetR domain, ChIP experiments showed a
24 hours after tetracycline addition in a clr4+-dependent manner. H3K9me2 ChIP-qPCR and H3K9me3
complete loss of the TetR occupancy signal at
ChIP-seq data for samples shown here are presented in fig. S2. Chromosome 3 (site of insertion of 10XtetO-
the 10XtetO-ade6+ locus (Fig. 5D). On the other
ade6+) and chromosome 1 (cen1L) coordinates are shown above the tracks, and read numbers (per
hand, we detected high levels of H3K9 dimethyl-
million) are indicated on the right. H3K9me2 at DNA repeats of cen1L serves as an internal control for
ation at the 10XtetO-ade6+ locus in red but not
the ChIP-seq data.
white isolates (Fig. 5E). We conclude that, once
SCIENCE sciencemag.org
3 APRIL 2015 • VOL 348 ISSUE 6230
RESEARCH RESEARCH ARTICLE
assembled, silent chromatin and histone H3K9me
hypothesis, the introduction of an additional
ment pathways, is primarily responsible for
at the 10XtetO-ade6+ locus can be maintained in
copy of WT clr4+, but not the clr4-W31G mutant,
RNAi-independent silencing in these deletion
the complete absence of the sequence-specific
into ago1D cells boosted the residual H3K9me
recruitment. To determine whether the initiator-
levels approximately threefold (Fig. 5F). Togeth-
independent silent state could also be inherited
er, these results support a direct "read-write"
through meiosis, we crossed red haploid cells of
mechanism in which Clr4 binds to preexisting
Our findings on mitotic and meiotic inheritance
opposite mating type, which lacked the TetR DNA
H3K9-methylated nucleosomes and catalyzes the
of H3K9me and silent chromatin in the absence
binding domain (clr4-ID, clr4+, epe1D), to obtain
methylation of H3K9 on newly deposited nucleo-
of sequence-specific recruitment—and without
diploid cells (Fig. 6A). These diploid cells were
somes to maintain heterochromatin independent-
requirement for a small RNA (sRNA) positive-
then sporulated, and after tetrad dissection, the
ly of the initial signals that induced methylation.
feedback loop associated with RNAi, or other
resulting haploid progeny were plated on low-
It is noteworthy that, in RNAi mutant cells, dele-
known modification systems such as DNA CpG
adenine medium. As shown in Fig. 6B, the result-
tions of epe1+, mst2+, and poz1+ boost H3K9me
methylation—strongly suggest that histones modi-
ing haploid cells formed mostly red or sectored
levels and restore silencing to varying degrees
fied by H3K9me can act as carriers of epigenetic
colonies, indicating that the silent state was also
at the pericentromeric repeats Fur-
information. This conclusion is supported by (i)
inherited through meiotic cell divisions.
thermore, consistent with its role in H3K9me
our demonstration that deletion of the fission
inheritance, the chromodomain of Clr4 is also
yeast putative histone H3K9 demethylase, epe1+,
Inheritance of H3K9me at
required for the RNAi-independent spreading
stabilizes the epigenetic OFF state and allows
native heterochromatin
of H3K9me at the mating type locus ). Our
its transmission through >50 cell divisions and
We next determined the extent to which epige-
results suggest that the epigenetic maintenance
(ii) the requirement for the Clr4 methyltrans-
netic maintenance mechanisms akin to what is
of H3K9me, rather than alternative establish-
ferase chromodomain, a domain that recognizes
observed for the ectopic locus might also operateat native S. pombe pericentromeric repeats. Al-
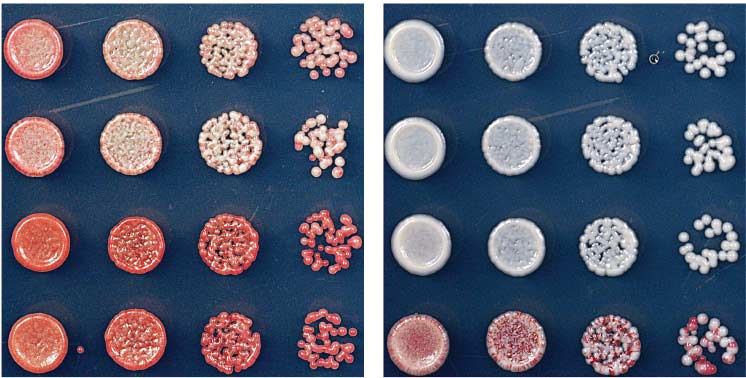
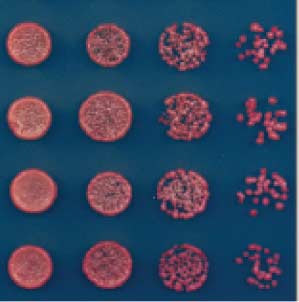
Fig. 3. Requirements
though RNAi is required for silencing of reporter
for maintenance of the
genes that are inserted within pericentromeric
repeat regions, deletion of RNAi components does
silent state. (A)
not entirely eliminate H3K9me and silencing
Establishment and
(Fig. 7A). Residual H3K9me in RNAi de-
maintenance require
letions might arise from either epigenetic mainte-
HP1 proteins (Swi6
nance mechanisms or weak RNAi-independent
and Chp2) and histone
establishment signals within centromeres
deacetylases (Clr3 and
To test for the presence of RNAi-independent sig-
Sir2). (B) Maintenance
nals that may operate at pericentromeric repeats,
of ectopic silencing in
we determined whether H3K9me could be es-
epe1D cells does not
tablished de novo at the repeats in cells with de-
require Dicer (Dcr1), as
letions of RNAi factors dcr1+ or ago1+. To perform
indicated by the growth
this experiment, we reintroduced clr4+ into clr4D,
D cells on
clr4D ago1D, or clr4D dcr1D cells (Fig. 7B) and used
ChIP-seq and ChIP-qPCR to quantify H3K9 di-
methylation levels. As shown in Fig. 7C, the re-
ectopic silencing in
introduction of clr4+ into clr4D cells fully restored
∆ dcr1 TetR-clr4-I, clr4+
epe1D cells does not
H3K9 dimethylation at the pericentromeric dg
require Argonaute
and dh repeats of chromosome 1. In contrast, clr4+
(Ago1), as indicated by
reintroduction into clr4D ago1D or clr4D dcr1D
the growth of red ago1D
double-mutant cells failed to promote any H3K9
cells on +tet medium.
dimethylation (Fig. 7, C and D). These results
(D) Either deleting epe1
show that RNAi is the primary mechanism for
(epe1D) or mutations
sequence-specific establishment of pericentro-
in its active site (epe1-
meric H3K9me domains and suggest that the
K314A or epe1-H297A)
H3K9me observed at these repeats after deletion
allow maintenance of the
of RNAi components results from epigenetic
off state after release of
the TetR-Clr4-I initiator.
Our ectopic heterochromatin experiments es-
(E) Replacement of
tablished a role for the chromodomain of Clr4
clr4+ with clr4DCD,
in epigenetic inheritance of H3K9me (Figs. 2 to
encoding Clr4 lacking
5). Consistent with the hypothesis that RNAi-
the chromodomain,
independent H3K9me at pericentromeric re-
abolishes initiator-
peats is maintained by epigenetic mechanisms,
residual H3K9me at the centromeric dg repeats
was abolished in ago1D clr4-W31G double-mutantcells (Fig. 7E). Clr4-W31G contains a mutation inthe chromodomain that attenuates binding to
H3K9me . The complete loss of H3K9me inago1D clr4-W31G double mutant cells therefore
TetR-clr4-I, clr4+, epe1∆
suggests that the residual H3K9me marks are
maintained by a mechanism that involves directchromodomain-dependent recruitment of Clr4
to preexisting marks. In further support of this
3 APRIL 2015 • VOL 348 ISSUE 6230
sciencemag.org SCIENCE
RESEARCH RESEARCH ARTICLE
di- and trimethylated H3K9, suggesting a direct
sequences or modulation of H3K9 demethyl-
raising the possibility that a posttranslational
read-write mechanism for this mode of epige-
ase activity in mammalian cells. Also, in fission
modification or cofactor may be required for
netic inheritance.
yeast, a previous study reported that ectopic
reconstitution of Epe1 demethylase activity.
Several recent studies have described the trans-
heterochromatin-dependent silencing of an ade6+
Regulation of histone demethylation activity may
generational inheritance of environmentally in-
allele induced by a centromeric DNA fragment
play a broad role in determining the reversibil-
duced changes in gene expression from parent to
is maintained in an RNAi-dependent manner
ity of epigenetic states. Although it is unknown
offspring ). The mechanism of this transgene-
after excision of the centromeric DNA fragment
whether Epe1 activity or levels regulate epige-
rational inheritance has not been fully defined
(suggesting that a sRNA amplification loop
netic transitions in S. pombe, regulation of his-
but appears to occur via both sRNA-dependent
may somehow be established at the ade6+ locus,
tone demethylase activity has been implicated in
and -independent pathways. In Caenorhabditis
which helps to maintain the silent state. How-
control of developmental transitions in multicel-
elegans, plants, fission yeast, and possibly other
ever, these findings contradict other studies, which
lular eukaryotes. In mouse embryonic stem cells,
systems, the transmission of histone modifica-
have demonstrated that silencing of ura4+ alleles
the pluripotency transcription factor Oct4 acti-
tion patterns is often coupled to sRNA generation
by the generation of sRNA from a hairpin could
vates the expression of Jumonji domain Jmjd1a
and/or CpG DNA methylation The latter
not be maintained in the absence of the inducing
and Jmjd2c H3K9 demethylases, and this activa-
pathways can form positive-feedback loops that
hairpin Our findings indicate that even in
tion appears to be important for stem cell self-
help maintain histone modification patterns
the absence of coupling to other positive-feedback
renewal An attractive possibility is that as
This coupling of positive-feedback loops would
loops, or in the absence of sequence-dependent
epigenetic states become established during tran-
increase the rate of reestablishment of silent do-
initiation signals, H3K9me defines a silent state
sition from pluripotency to the differentiated state,
mains and thus counteract the erasure activity
that can be epigenetically inherited (fig. S7). Main-
reduction in the expression of H3K9 demethyl-
of enzymes such as Epe1 or mechanisms that in-
tenance of the OFF state is probably determined by
ases helps to stabilize the differentiated state
crease the rate of histone turnover.
the balance between the rate of H3K9me by the
In another example, down-regulation of the amine
A previous study used a small-molecule dimer-
Clr4 reader-writer module and the loss rate due to
oxidase family histone demethylase LSD1 during
ization strategy to show that ectopically induced
demethylation by an Epe1-dependent mechanism,
activation of individual olfactory receptor (OR)
domains of H3K9me at the Oct4 locus in murine
transcription-coupled nucleosome exchange, and
genes in the mammalian nose has been suggested
fibroblasts can be maintained after the removal of
dilution of histones during DNA replication.
to create an epigenetic "trap" that prevents the
the small-molecule inducer by a mechanism that
Although demethylase activity for the S. pombe
activation of additional OR genes (). More gen-
is reinforced by CpG DNA methylation . How-
Epe1 protein has not yet been demonstrated
erally, H3K9 demethylases may act as surveillance
ever, unlike the experiments presented here, the
in vitro, key residues required for demethylase
enzymes that prevent the formation of spurious
use of the Oct4 locus, which is normally packaged
activity in other Jumonji domain proteins are
H3K9 methylated domains, which may lead to
into heterochromatin in fibroblasts , precludes
conserved in Epe1 and required for its in vivo
epigenetic mutations and gene inactivation.
any conclusions about sequence-independent in-
effects on silencing (, and its effect on
heritance, as contributions from locus-specific se-
initiator-independent inheritance of heterochro-
Materials and methods
quence elements that normally silence Oct4 in
matin (this study, Fig. 4C). In addition, a recent
differentiated cells cannot be ruled out. It there-
study showed that the activity of human PHF2,
Plasmids containing 10XtetO binding sites up-
fore remains to be determined whether H3K9me
another member of the Jumonji domain protein
stream of ade6+ and ura4-GFP reporter genes
can be inherited independently of specific DNA
family, is regulated by phosphorylation (),
were constructed by first synthesizing a plasmid
100 hours
TetR-clr4-I, clr4+, epe1+
TetR-clr4-I, clr4+, epe1∆
Fig. 4. Kinetics of decay of the silent state after
release of TetR-Clr4-I using a GFP reporter gene
reveals epigenetic maintenance in epe1+ cells.
(A) Schematic diagram of the 10XtetO-ura4-GFP
locus. (B to E) FACS analysis of GFP expressionin the indicated strains at 0, 6, 23, and 100 hours
after tetracycline addition shows the time evolu-
tion of the distribution of GFP-OFF cells. a.u., ar-
bitrary units. (F) Data for time points between
0 and 100 hours after addition of tetracycline
were plotted to display the fraction of GFP-OFF
Fluorescence (a.u.)
cells as a function of time. Dose-response curvefitting was used as a guide.
SCIENCE sciencemag.org
3 APRIL 2015 • VOL 348 ISSUE 6230
RESEARCH RESEARCH ARTICLE
containing 10 tetO sites flanked by 200-bphomology sequences to facilitate reporter in-sertion at the ura4 locus. The reporter genes werecloned downstream of the tetO binding sites
using PacI and AscI restriction sites that were
incorporated during the initial synthesis of the
deletion of TetR DNA
plasmid. The ade6+ reporter construct consists of
the full-length WT ade6+ gene with endogenous
FLAG ChIP
upstream promoter and downstream terminator
sequences. The ura4-GFP reporter consists of thefull-length ura4+ gene fused at the C terminuswith a monomeric yeast codon–optimized GFP
using Gibson assembly (This construct was
epe1∆, red isolate
subsequently cloned downstream of the 10 tetO
sites and appended with a 114-bp ura4 promoterelement and the corresponding endogenous ura4
downstream terminator sequence. The plasmid
containing TetR-clr4-I was constructed by modify-ing a pFA6a-natMX6-P
H3K9me2 ChIP
nmt1 plasmid. The promoter
elements in the original plasmid were replacedwith the endogenous clr4+ promoter (using BglIIand PacI restriction sites). The TetR construct con-
sists of an N-terminal SV40 nuclear localization
sequence followed immediately by a 2X-FLAG
epe1∆, white isolate
tag. The clr4+ chromodomain deletion construct
consists of a clr4 allele lacking amino acids 7 to59. The synthesis of the TetR-clr4-I fusion with the
upstream endogenous clr4 promoter elements
was achieved by Gibson assembly. The deletion
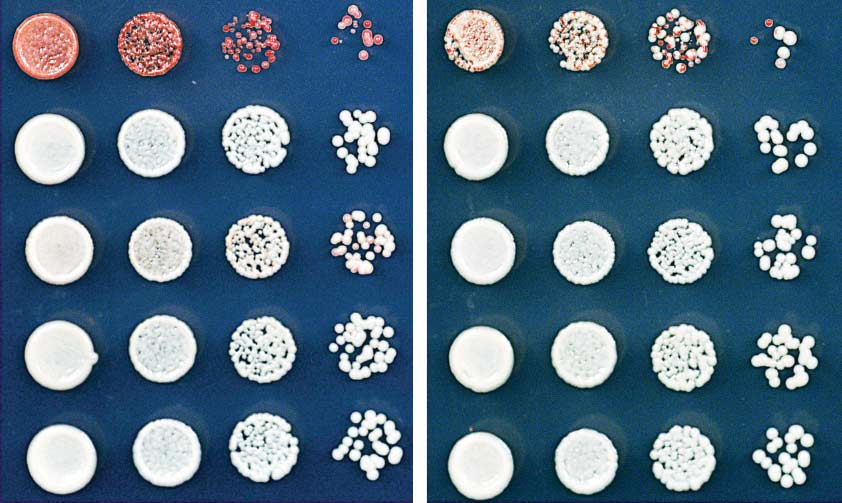
Fig. 5. Silencing and H3K9me are maintained after deletion of the
of the TetR DNA binding element was achieved
TetR DNA binding domain. (A) Diagram showing the experimental
after modifying a pFA6a-hphMX6-P
,
scheme for conversion of TetR-clr4-I to clr4-ID, which lacks recruitment
by insertion of the endogenous clr4+ promoter
activity because it can no longer bind to tetO sites. The isolation of sec-
,
red isolate
and a clr4 allele lacking the chromodomain.
tored and red clr4-I
D cells demonstrates maintenance in the complete
absence of the TetR DNA binding domain (B), whereas white clr4-ID
colonies indicate irreversible loss of silencing (C). (D) ChIP-qPCR ex-
A strain containing the 10 tetO sites was first
periments show that FLAG-tagged TetR-Clr4 signal could be detected
made by insertion of the reporter gene at the
at tetO sites in clr4-ID cells. (E) ChIP-qPCR experiments show elevated
ura4+ locus. The subsequent introduction of the
levels of H3K9me in silent clr4-ID cells (red isolates) but background
TetR-Clr4-I fusion protein was achieved with the use
levels of methylation in ade6+-expressing (white) clr4-ID cells. Error
of a PCR-based gene-targeting approach ().
bars in (D) and (E) represent SD.
Strains with the designation TetR-Clr4-I are thosein which the endogenous copy of clr4 is replacedwith the TetR-Clr4-I fusion, making it the only
Fig. 6. Inheritance of initiator-
source of Clr4 expression in the cells. In strains
independent silencing through meiotic
where the WT copy of clr4+ is intact (i.e., TetR-
cell divisions. (A) Scheme for mating of
clr4-I,clr4+), the fusion protein is inserted at the
silent (red) ade6+ haploid cells of the
trp1+ locus. The deletions of the various RNAi and
indicated genotypes in which the TetR-
chromatin components were achieved either by
Clr4-I was deleted. After sporulation of
PCR-based gene-targeting approaches or by a
the resulting diploid cells, tetrads were
cross followed by random spore analysis and
dissected, and the haploid meiotic prog-
PCR-based screening to select for colonies that
eny were plated on low-adenine medium
harbored the reporter gene, the TetR fusion pro-
(B). Results of four tetrad dissections are
tein, and the appropriate deletion. Strains con-
presented and show inheritance and
taining deletions of the TetR DNA binding domain
variegation of the silent state.
(clr4-ID) were constructed by both PCR-based tar-geting approaches and crosses followed by ran-dom spore analysis. The resulting colonies weretested using allele specific primers. To isolate redcolonies that harbor a deletion of the TetR DNAbinding domain, sectored colonies, which testedpositive for the deletion in the allele-specific PCRscreen, were replated to isolate single red colonieson plates containing limiting adenine. All strainsused in this study are listed in table S1.
Crosses were performed between red isolates
of haploid cells of opposite mating type thatharbored a deletion of the TetR DNA binding
3 APRIL 2015 • VOL 348 ISSUE 6230
sciencemag.org SCIENCE
RESEARCH RESEARCH ARTICLE
module. The resulting diploid, which lacks any
30 min at room temperature before quenching
sequence-specific establishment factors, was then
Cells were grown to a density of 2.5 × 107 cells/ml at
with 125 mM glycine for 5 min. The subsequent
allowed to sporulate. After tetrad dissection, spores
32°C in yeast extract supplemented with adenine
steps for sample processing were performed as
were plated on low-adenine medium and allowed
(YEA) or YEA containing tetracycline (2.5 mg/ml).
previously described (). Immunoprecipitation
to grow at 32°C for 3 days.
Cells were cross-linked with 1% formaldehyde for
was performed using the following antibodies:2.5 ml a-H3K9me2 (ab1220, Abcam) for quanti-fying H3K9me2 levels, 2 mg a-H3K9me3 ) forquantifying H3K9me3 levels, and 2.5 ml a-FLAG(M2, Sigma) for quantifying TetR-Clr4-I occupan-cy at the ectopic locus before and after additionof tetracycline. DNA purified from the ChIP ex-periments was analyzed by quantitative PCR usingan Applied Biosystems 7900HT Fast Real-TimePCR system. See table S2 for primer sequences.
ChIP-seq libraries were constructed, sequencedusing an Illumina HiSeq platform, and processedas described previously ).
Strains containing the ade6+ reporter constructwere grown overnight, after which fivefold dilu-tions of each culture were spotted on plates con-taining only yeast extract and glucose withoutany additional adenine supplements with (+tet)or without (–tet) tetracycline (2.5 mg/ml). Eachsilencing assay also included centromeric silenc-ing reporter strains that are unresponsive to tetra-cycline (otr1R:ade6+ and ura4::10XtetO- ade6+) ascontrols to ensure that the addition of tetracyclinedoes not induce any changes in reporter geneexpression.
Cells containing TetR-clr4-I and 10XtetO- ura4-GFP reporter were maintained in log phase ( 2.5 ×107 cells/ml) through the course of sample prep-aration at various time points after addition oftetracycline (2.5mg/ml). Approximately 2.5 × 107cells were harvested and fixed by addition of 70%ethanol for 20 min. The cells were then washedtwice with 1X tris-buffered saline (TBS) (200 mMTris pH 7.5, 150 mM NaCl) and resuspendedin 1 ml of 1X TBS in a FACS tube (BD Falcon).
GFP fluorescence was then measured using aFACScalibur instrument (Becton Dickinson), andexcitation was achieved by using an argon laseremission of 488 nm. Data collection was performedusing Cellquest software (Becton Dickinson), anda primary gate based on physical parameters(forward and side light scatter) was set to ex-clude dead cells or debris. Typically, 20,000 cellswere analyzed for each sample and time point.
The resulting GFP fluorescence profiles were fit
Fig. 7. RNAi-independent H3K9me at pericentromeric repeats is epigenetically inherited. (A)
using Gaussian curves (Origin 8.0), assuming a
ChIP-seq experiments showing the persistence of residual histone H3K9me2 at the pericentromeric
model in which cells exhibit two expression states:
dg and dh repeats of chromosome 1 in ago1D and dcr1D cells. Libraries were sequenced on the Illumina
either GFP-ON or GFP-OFF. The fraction of cells
HiSeq2500 platform and normalized to reads per million (y axis). Chromosome coordinates are indicated
in each state was calculated by measuring the
above the plots. (B) Scheme for the reintroduction of clr4+ into RNAiD, clr4D cells to test the requirement for
area under the curve for each Gaussian fit.
RNAi in H3K9me establishment. clr4+ was reintroduced to the native locus to avoid overexpression. (C)
ChIP-seq experiments showing that the reintroduction of clr4+ into clr4D cells, but not clr4D ago1D or clr4D
L. Ringrose, R. Paro, Epigenetic regulation of cellular memory
dcr1D cells, restores H3K9me2 at the pericentromeric repeats of chromosome 1 (left). Reads for H3K9me2
by the Polycomb and Trithorax group proteins. Annu. Rev. Genet.
at the telomeres of chromosome 1 (telL1) on the right side show that, unlike the centromeres, establishment
38, 413–443 (2004). doi:
of telomeric H3K9me does not require RNAi. (D) ChIP-qPCR experiments verify that RNAi is required for the
reestablishment of H3K9me2 at the pericentromeric dg repeats. (E) ChIP-qPCR experiments show that a
2. D. Moazed, Mechanisms for the inheritance of chromatin
states. Cell 146, 510
mutation in the chromodomain of Clr4 (clr4W31G) abolishes the maintenance of H3K9me2 at dg repeats.
–518 (2011). doi:
(F) ChIP-qPCR experiments show that an additional copy of WT clr4+, but not clr4W31G, boosts residual
3. K. Luger, A. W. Mäder, R. K. Richmond, D. F. Sargent,
H3K9me2 levels at dg. Error bars in (D) to (F) represent SD.
T. J. Richmond, Crystal structure of the nucleosome core
SCIENCE sciencemag.org
3 APRIL 2015 • VOL 348 ISSUE 6230
RESEARCH RESEARCH ARTICLE
particle at 2.8 A resolution. Nature 389, 251–260 (1997).
telomeres in yeast. Cell 75, 531–541 (1993). doi:
Nat. Rev. Genet. 11, 204–220 (2010). doi:
4. T. Jenuwein, C. D. Allis, Translating the histone code. Science
26. A. Kagansky et al., Synthetic heterochromatin bypasses RNAi
46. N. A. Hathaway et al., Dynamics and memory of
293, 1074–1080 (2001). doi:
and centromeric repeats to establish functional centromeres.
heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Science 324, 1716–1719 (2009). doi:
5. B. Li, M. Carey, J. L. Workman, The role of chromatin during
47. B. S. Wheeler, B. T. Ruderman, H. F. Willard, K. C. Scott,
transcription. Cell 128, 707–719 (2007). doi:
27. M. Gossen, H. Bujard, Tight control of gene expression
Uncoupling of genomic and epigenetic signals in the
in mammalian cells by tetracycline-responsive promoters.
maintenance and inheritance of heterochromatin domains in
6. T. Kouzarides, Chromatin modifications and their function.
Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 (1992).
fission yeast. Genetics 190, 549–557 (2012). doi:
Cell 128, 693–705 (2007). doi: ;
28. B. D. Reddy et al., Elimination of a specific histone H3K14
48. T. Iida, J. Nakayama, D. Moazed, siRNA-mediated
7. S. L. Schreiber, B. E. Bernstein, Signaling network model of
acetyltransferase complex bypasses the RNAi pathway to
heterochromatin establishment requires HP1 and is associated
chromatin. Cell 111, 771–778 (2002). doi:
regulate pericentric heterochromatin functions. Genes Dev. 25,
with antisense transcription. Mol. Cell 31, 178–189 (2008).
214–219 (2011). doi: pmid:
8. O. J. Rando, Combinatorial complexity in chromatin structure and
29. X. Tadeo et al., Elimination of shelterin components bypasses
49. R. Yu, G. Jih, N. Iglesias, D. Moazed, Determinants of
function: Revisiting the histone code. Curr. Opin. Genet. Dev. 22,
RNAi for pericentric heterochromatin assembly. Genes Dev. 27,
heterochromatic siRNA biogenesis and function. Mol. Cell
148–155 (2012). doi: ; pmid:
2489–2499 (2013). doi: pmid:
53, 262–276 (2014). doi:
9. V. Jackson, R. Chalkley, Separation of newly synthesized
30. S. C. Trewick, E. Minc, R. Antonelli, T. Urano, R. C. Allshire,
nucleohistone by equilibrium centrifugation in cesium chloride.
The JmjC domain protein Epe1 prevents unregulated assembly
50. Y. Shi, J. R. Whetstine, Dynamic regulation of histone lysine
Biochemistry 13, 3952–3956 (1974). doi:
and disassembly of heterochromatin. EMBO J. 26, 4670–4682
methylation by demethylases. Mol. Cell 25, 1–14 (2007).
(2007). doi: ; pmid:
10. J. M. Sogo, H. Stahl, T. Kol er, R. Knippers, Structure of replicating
31. Y. Tsukada et al., Histone demethylation by a family of JmjC
51. A. Baba et al., PKA-dependent regulation of the histone
simian virus 40 minichromosomes. The replication fork, core
domain-containing proteins. Nature 439, 811–816 (2006).
lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 13,
histone segregation and terminal structures. J. Mol. Biol. 189,
668–675 (2011). doi: pmid:
189–204 (1986). doi: ;
32. E. B. Gómez, J. M. Espinosa, S. L. Forsburg, Schizosaccharomyces
52. Y. H. Loh, W. Zhang, X. Chen, J. George, H. H. Ng, Jmjd1a
pombe mst2+ encodes a MYST family histone acetyltransferase
and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal
11. M. Radman-Livaja et al., Patterns and mechanisms of
that negatively regulates telomere silencing. Mol. Cell. Biol. 25,
in embryonic stem cells. Genes Dev. 21, 2545–2557 (2007).
ancestral histone protein inheritance in budding yeast.
8887–8903 (2005). doi:
PLOS Biol. 9, e1001075 (2011). doi:
53. D. B. Lyons et al., An epigenetic trap stabilizes singular
33. A. Roguev et al., High conservation of the Set1/Rad6 axis of
olfactory receptor expression. Cell 154, 325–336 (2013).
12. A. V. Probst, E. Dunleavy, G. Almouzni, Epigenetic inheritance
histone 3 lysine 4 methylation in budding and fission yeasts.
during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206
J. Biol. Chem. 278, 8487–8493 (2003). doi:
54. D. G. Gibson et al., Enzymatic assembly of DNA molecules
(2009). doi: pmid:
up to several hundred kilobases. Nat. Methods 6, 343–345
13. B. D. Strahl, C. D. Allis, The language of covalent histone
34. T. Miyoshi, J. Kanoh, M. Saito, F. Ishikawa, Fission yeast
(2009). doi: pmid:
modifications. Nature 403, 41–45 (2000). doi:
Pot1-Tpp1 protects telomeres and regulates telomere length.
55. J. Bähler et al., Heterologous modules for efficient and
Science 320, 1341–1344 (2008). doi:
versatile PCR-based gene targeting in Schizosaccharomyces
14. B. M. Turner, Histone acetylation and an epigenetic code.
pombe. Yeast 14, 943–951 (1998). doi:
BioEssays 22, 836–845 (2000). doi:
35. T. A. Volpe et al., Regulation of heterochromatic silencing and
histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837
15. M. Ptashne, On the use of the word ‘epigenetic'. Curr. Biol.
(2002). doi: pmid:
56. E. D. Egan, C. R. Braun, S. P. Gygi, D. Moazed, Post-
17, R233–R236 (2007). doi: ;
36. M. Sadaie, T. Iida, T. Urano, J. Nakayama, A chromodomain
transcriptional regulation of meiotic genes by a nuclear RNA
protein, Chp1, is required for the establishment of
silencing complex. RNA 20, 867–881 (2014). doi:
16. R. Margueron, D. Reinberg, Chromatin structure and the
heterochromatin in fission yeast. EMBO J. 23, 3825–3835
inheritance of epigenetic information. Nat. Rev. Genet. 11,
(2004). doi: ; pmid:
57. T. Hattori et al., Recombinant antibodies to histone
285–296 (2010). doi: pmid:
37. M. Halic, D. Moazed, Dicer-independent primal RNAs trigger
post-translational modifications. Nat. Methods 10, 992–995
17. T. H. Cheng, M. R. Gartenberg, Yeast heterochromatin is a
RNAi and heterochromatin formation. Cell 140, 504–516
(2013). doi: pmid:
dynamic structure that requires silencers continuously.
(2010). doi: pmid:
Genes Dev. 14, 452–463 (2000). pmid:
38. F. E. Reyes-Turcu, K. Zhang, M. Zofall, E. Chen, S. I. Grewal,
18. S. G. Holmes, J. R. Broach, Silencers are required for
Defects in RNA quality control factors reveal RNAi-independent
We thank A. Shetty for help with tetrad dissections; D. Shea for
inheritance of the repressed state in yeast. Genes Dev. 10,
nucleation of heterochromatin. Nat. Struct. Mol. Biol. 18,
help in data processing; DNA 2.0 (Menlo Park, CA) for synthesis
1021–1032 (1996). doi: pmid:
1132–1138 (2011). doi: ; pmid:
of the plasmid containing tetO binding sites; D. Landgraf for
19. A. K. Sengupta, A. Kuhrs, J. Müller, General transcriptional
39. J. Nakayama, J. C. Rice, B. D. Strahl, C. D. Allis, S. I. Grewal,
the gift of a yeast codon–optimized GFP plasmid; T. Hattori and
silencing by a Polycomb response element in Drosophila.
Role of histone H3 lysine 9 methylation in epigenetic control
S. Koide for the gift of a H3K9me3 recombinant antibody; and
Development 131, 1959–1965 (2004). doi:
of heterochromatin assembly. Science 292, 110–113 (2001).
members of the Moazed lab for helpful discussions and comments—
particularly E. Egan, N. Iglesias, and J. Xiol for providing
20. A. Busturia, C. D. Wightman, S. Sakonju, A silencer is required
40. K. Zhang, K. Mosch, W. Fischle, S. I. Grewal, Roles of the Clr4
valuable experimental help and advice and R. Behrouzi for advice
for maintenance of transcriptional repression throughout
methyltransferase complex in nucleation, spreading and
and help with FACS analysis. The raw and processed ChIP-seq
Drosophila development. Development 124, 4343–4350 (1997).
maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15,
data are publicly available at the National Center for Biotechnology
381–388 (2008). doi: pmid:
Information Gene Expression Omnibus under accession number
21. M. Bühler, S. M. Gasser, Silent chromatin at the middle and
41. E. Heard, R. A. Martienssen, Transgenerational epigenetic
GSE63304. This work was supported by a postdoctoral
ends: Lessons from yeasts. EMBO J. 28, 2149–2161 (2009).
inheritance: Myths and mechanisms. Cell 157, 95–109 (2014).
fellowship from the Leukemia and Lymphoma Society
(CDP-5025-14 to K.R.) and a grant from the NIH (GM072805 to
22. R. C. Allshire, J. P. Javerzat, N. J. Redhead, G. Cranston, Position
42. S. G. Gu et al., Amplification of siRNA in Caenorhabditis elegans
D.M.). G.J. was supported partly by an NIH training grant
effect variegation at fission yeast centromeres. Cell 76, 157–169
generates a transgenerational sequence-targeted histone
(T32 GM007226). D.M. is an investigator of the Howard Hughes
(1994). doi: ; pmid:
H3 lysine 9 methylation footprint. Nat. Genet. 44, 157–164
Medical Institute.
23. S. I. Grewal, A. J. Klar, Chromosomal inheritance of epigenetic
(2012). doi: pmid:
states in fission yeast during mitosis and meiosis. Cell 86,
43. X. Zhong et al., Molecular mechanism of action of plant DRM
SUPPLEMENTARY MATERIALS
95–101 (1996). doi:
de novo DNA methyltransferases. Cell 157, 1050–1060 (2014).
24. J. Nakayama, A. J. Klar, S. I. Grewal, A chromodomain protein,
44. M. R. Motamedi et al., Two RNAi complexes, RITS and RDRC,
Swi6, performs imprinting functions in fission yeast during
physically interact and localize to noncoding centromeric
mitosis and meiosis. Cell 101, 307–317 (2000). doi:
RNAs. Cell 119, 789–802 (2004). doi:
14 July 2014; accepted 13 November 2014
25. C. T. Chien, S. Buck, R. Sternglanz, D. Shore, Targeting of SIR1
45. J. A. Law, S. E. Jacobsen, Establishing, maintaining and
Published online 20 November 2014;
protein establishes transcriptional silencing at HM loci and
modifying DNA methylation patterns in plants and animals.
3 APRIL 2015 • VOL 348 ISSUE 6230
sciencemag.org SCIENCE
Source: http://www.dbt.univr.it/documenti/OccorrenzaIns/matdid/matdid529668.pdf
What GlaxoSmithKline Does Not Seem to Want Anyone to Know About Paxil Individuals of al ages should be closely monitored for suicidal and homicidal thoughts and behaviours for at least 1-month after they start taking Paxil and after they increase their dosage. They should also be closely monitored after they stop taking Paxil. By David Carmichael
Les anticancéreux oraux Un regard différent N°8 Septembre 2014 Avec ce nouveau numéro d'Onco News, toute la CAHPP se joint à moi pour vous souhaiter une excellente rentrée 2014. Dans cette lettre en Cancérologie, la CAHPP réaffirme son rôle de centrale de conseil et de référencement souhaitant apporter son expertise en matière d'optimisation des achats et des








