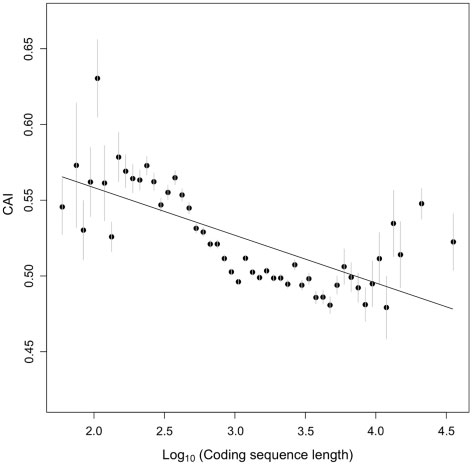
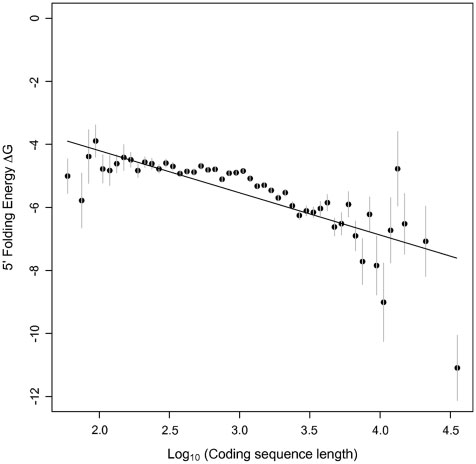
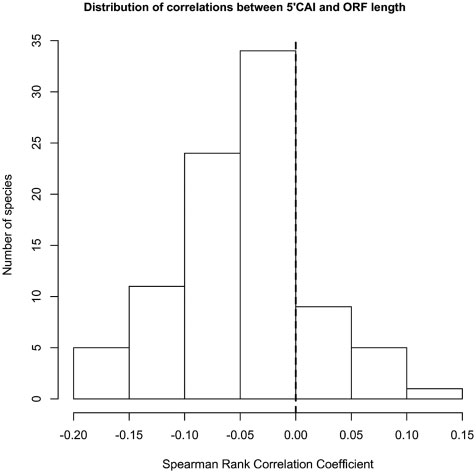
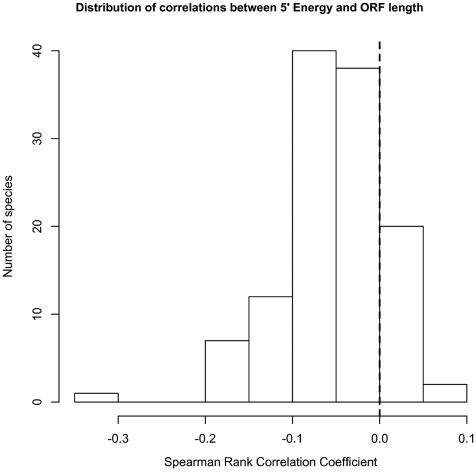
Percutaneous aortic valve replacement: overview and suggestions for anesthestic management
Journal of Clinical Anesthesia (2010) 22, 373–378 Percutaneous aortic valve replacement: overview andsuggestions for anesthestic management☆ Hermann Heinze MD (Assistant Professor)a,⁎, Holger Sier MD, (Staff Cardiac Surgeon)b,Ulrich Schäfer MD (Senior Cardiologist)c, Matthias Heringlake MD, PhD (Professor)a aDepartment of Anesthesiology, University of Lübeck, 23538 Lübeck, GermanybDepartment of Cardiothoracic Surgery, University of Lübeck, 23538 Lübeck, GermanycDepartment of Cardiology, Asklepios Klinik St. Georg, 20099 Hamburg, Germany