Www-personal.ksu.edu

Lack of Association of the S769N Mutation in Plasmodium falciparum
SERCA (PfATP6) with Resistance to Artemisinins
Long Cui,a Zenglei Wang,a Hongying Jiang,b Daniel Parker,a Haiyan Wang,c Xin-Zhuan Su,b and Liwang Cuia
Department of Entomology, The Pennsylvania State University, University Park, Pennsylvania, USAa; Laboratory of Malaria and Vector Research, National Institute of Allergy
and Infectious Diseases, National Institutes of Health, Bethesda, Maryland,USAb; and Department of Statistics, Kansas State University, Manhattan, Kansas, USAc
The recent emergence of artemisinin (ART) resistance in Plasmodium falciparum in western Cambodia, manifested as delayed
parasite clearance, is a big threat to the long-term efficacy of this family of antimalarial drugs. Among the multiple candidate
genes associated with ART resistance in P. falciparum, the sarcoplasmic/endoplasmic reticulum Ca2ⴙ-ATPase PfATP6 has been
postulated as a specific target of ARTs. The PfATP6 gene harbors multiple single-nucleotide polymorphisms in field parasite
populations, and S769N has been associated with decreased sensitivity to artemether in parasite populations from French Gui-
ana. In this study, we used an allelic exchange strategy to engineer parasite lines carrying the S769N mutations in P. falciparum
strain 3D7 and evaluated whether introduction of this mutation modulated parasite sensitivity to ART derivatives. Using three
transgenic lines carrying the 769N mutation and two transgenic lines carrying the wild-type 769S as controls, we found that
S769N did not affect PfATP6 gene expression. We compared the sensitivities of these parasite lines to three ART derivatives, arte-
mether, artesunate, and dihydroartemisinin, in 18 biological experiments and detected no significant effect of the S769N muta-
tion on parasite response to these ART derivatives. This study provides further evidence for the lack of association of PfATP6
with ART resistance.
Artemisinin (ART) and its derivatives play an indispensable inducedtemporaryarrestofgrowth(dormancy)atthisstage
role in the malaria elimination/eradication campaigns cur-
Whereas this may partially explain the prolonged parasite
rently being unfolded in many regions where malaria is endemic.
clearance observed in clinical studies the possibility of host
To reduce the chance of resistance development and prolong the
factors that may play a crucial role in determining prolonged par-
life span of this group of drugs, the World Health Organization
asite clearance times observed in vivo has not been investigated
(WHO) has endorsed ART-based combination therapies (ACTs)
In addition, it has been proposed that ARTs may interfere
as the first-line treatment for Plasmodium falciparum malaria
with the mitochondrial function of the parasite Other
Since the adoption of the ACT policy in many regions where P.
postulated cellular targets of ARTs include the multidrug resis-
falciparum malaria is endemic a trend of steady reduction in
tance 1 (mdr1) gene, ABC transporter genes G7 and G49
global malaria incidence has been observed However, the
translationally controlled tumor protein and the sarcoplas-
recent detection of emerging low-grade resistance to ARTs in
mic/endoplasmic reticulum Ca2⫹-ATPase (SERCA) ortholog
western Cambodia, manifested as delayed parasite clearance, has
PfATP6 In rodent malaria caused by Plasmodium chabaudi, a
raised a major concern The Greater Mekong Subregion
mutation in the deubiquitinating enzyme ubp-1 has been mapped
(GMS) has been an epicenter of drug resistance, and resistance to
as a determinant of experimentally selected ART resistance
chloroquine (CQ) and pyrimethamine has spread from there to
Despite these proposed targets, no definite genetic determinant of
Africa Therefore, an analogous spread of ART resistance
Plasmodium sensitivity to ARTs has been identified so far. More-
from this region would be a disaster. As WHO has been gathering
over, none of these candidate genes appears to be responsible for
resources for eliminating and containing ART-resistant parasites
the observed ART resistance in western Cambodia
surveillance efforts have intensified in the GMS, where ART
The proposal of PfATP6 as the primary target of ARTs in ma-
use has the longest history. Meanwhile, research aimed to deci-
laria parasites was initially based on the structural resemblance of
pher the underlying mechanisms of ART resistance has become a
ARTs to thapsigargin, a specific inhibitor of mammalian SERCAs.
Since PfATP6 is the only SERCA-type Ca2⫹-ATPase in the malaria
ARTs contain an endoperoxide bridge that is essential for the
parasite's genome, it was evaluated as the target of ARTs. When
parasite-killing activities Although the structure of ART was
expressed in Xenopus laevis oocytes, PfATP6 can be specifically
solved over 3 decades ago, the mode of action of this group of
inhibited by ART as well as thapsigargin Modeling of
drugs has not been unequivocally determined The
PfATP6 and docking simulations suggest that ARTs bind to
most-studied model suggests that heme-mediated activation ofARTs results in C-centered free radicals that alkylate biomoleculesin the parasite, leading to parasite death Evidence
Received 13 October 2011 Returned for modification 5 December 2011
supporting the involvement of heme in the action of ARTs in-
Accepted 9 February 2012
cludes antagonistic actions of iron chelators and the requirement
Published ahead of print 21 February 2012
of hemoglobin digestion for the activity of ART This also
Address correspondence to Liwang Cui, [email protected].
correlates with the tolerance phenomenon of ring-stage parasites
Copyright 2012, American Society for Microbiology. All Rights Reserved.
to ARTs, when hemoglobin digestion activity is low. The reduced
metabolic activity at the ring stage is reflected further in ART-
Antimicrobial Agents and Chemotherapy
PfATP6 and Artemisinin Resistance in P. falciparum
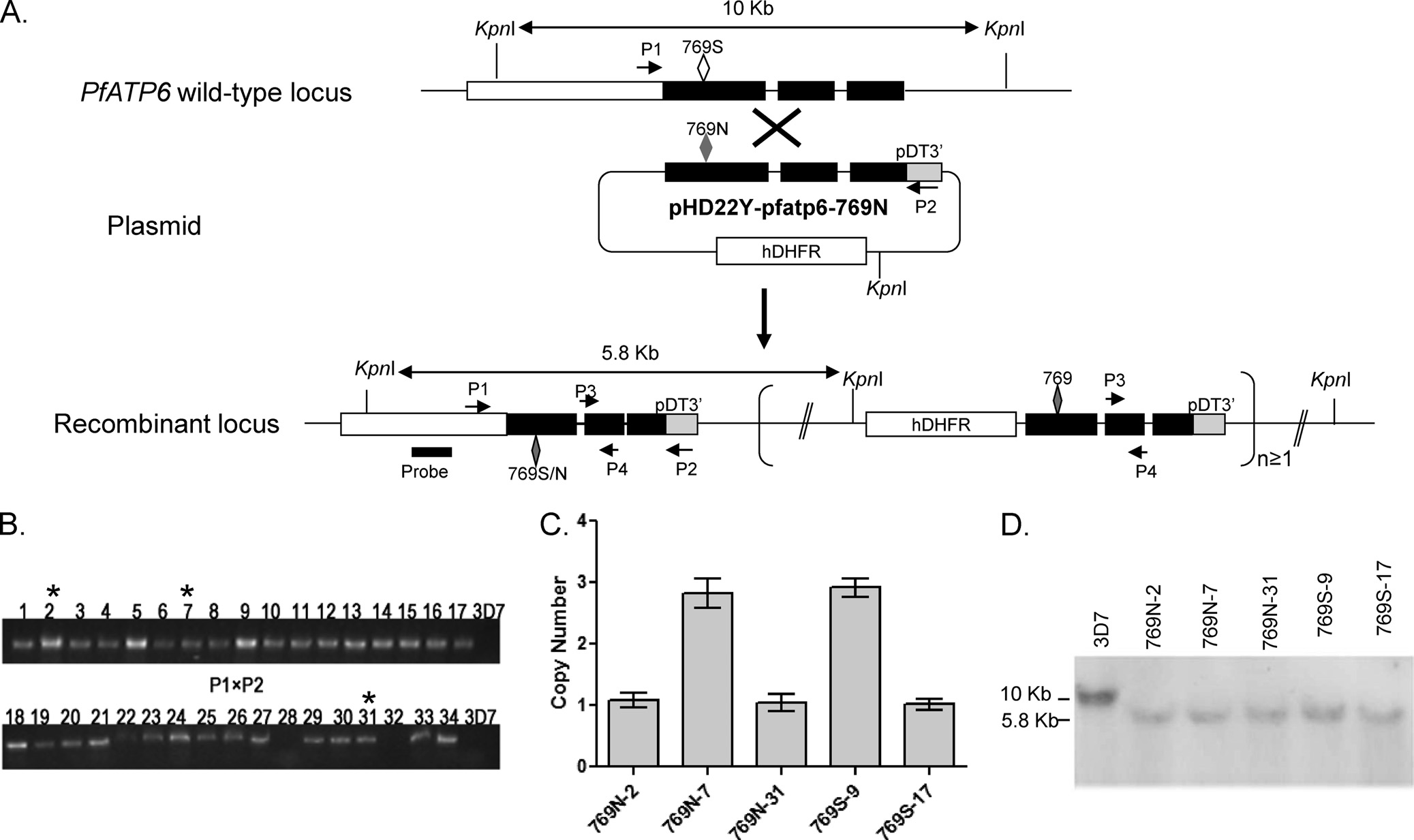
FIG 1 Development of transgenic lines in 3D7 with the PfATP6 S769N mutation. (A) Schematic representation of single-crossover event at the Pfatp6 locus.
(Top) The Pfatp6 locus on chromosome 1. Solid lines represent introns or intergenic regions, and filled boxes indicate the coding regions. (Middle) Plasmid
pHD22y-pfatp6-769N, showing the Pfatp6 genomic fragment and the drug selection cassette hDHFR. (Bottom) Predicted single-crossover events at the Pfatp6
locus. The fragment within the bracket indicates the scenario when integration of concatemerized plasmid occurs. The C-terminal fragment of PfATP6 cloned
in the transfection plasmid is shown as filled boxes. The open and filled lozenges indicate the locations of the wild-type and mutant amino acids at position 769,
respectively. Restriction enzyme KpnI sites and the expected sizes of DNA fragments after KpnI digestion are illustrated. The positions and orientations of the
primers on chromosome 1 and the plasmid are shown. Primer pairs P1 and P2 were used for integration-specific PCR, whereas primers P3 and P4 were used for
determining the copy number of the integrated plasmid. The position of the probe used for genomic Southern blotting is also marked. (B) Integration-specific
PCR products, based on used of primer P1 on chromosome 1 and primer P2 on the plasmid, showing 32 positive and 2 negative clones (lanes 28 and 32). The PCR
products of 32 positive clones were sequenced, and asterisks indicate the three clones with the S769N mutation. (C) Copy numbers of the integrated plasmid or
concatemers at the Pfatp6 locus as determined by real-time PCR using primer pairs P3 and P4. Shown here are five transgenic clones, with two containing the
wild-type residue (739S-9 and 769S-17) and three with the 769N mutation (769N-2, 769N-7, and 769N-31). (D) Genomic Southern blot of DNA isolated from
3D7 and five clones with plasmid integration at the Pfatp6 locus. Genomic DNA was digested with KpnI and separated in a 1% agarose gel. The blot was
hybridized with the probe marked in panel A, which revealed a ca. 10-kb fragment in 3D7 and 5.8-kb fragment in the recombinant Pfatp6 locus.
PfATP6 through hydrophobic interactions Variations at
position 263 has not been detected in field isolates from regions
a single residue, 263, located in the predicted ART-binding pocket
with suspected ART resistance, Jambou et al. reported an associ-
of PfATP6, tremendously affect the sensitivity of the enzyme to
ation of reduced in vitro ATM susceptibility with an S769N sub-
ARTs When assayed in X. laevis oocytes, the introduction of
stitution in a limited number of parasite field isolates from French
a single substitution, L263A or L263S (residues in Plasmodium
Guiana Additionally, this mutation was later detected in a
vivax and Plasmodium berghei SERCAs, respectively) resulted in
few isolates from Senegal, and it was associated with higher IC50s
an approximately 3-fold increase or decrease of sensitivity to
for artesunate (ATS) Whereas this substitution was consid-
ARTs, respectively. Furthermore, the L263E replacement led to
ered rare in previous analyses a recent study of
complete abolishment of inhibition by ART However, this
parasite isolates obtained from travelers to Africa suggested that it
observation was not extended to P. falciparum, where introduc-
might be quite prevalent in Africa However, an in vitro anal-
tion of the L263E mutation through transgenics resulted in bor-
ysis of a single African isolate carrying the S769N mutation
derline nonsignificant changes in the 50% inhibitory concentra-
showed sensitivity to dihydroartemisinin (DHA) and ATM
tions (IC50s) for ART and its derivatives Recently, Lepore et
Thus, the role of the S769N mutation of PfATP6 in resistance
al. performed modeling and docking simulations for SERCA pro-
to ARTs remains to be verified experimentally.
teins from P. falciparum, Schistosoma mansoni, and humans, but
To elucidate a possible role of PfATP6 in ART resistance, we
they did not find significant differences in the binding mode of
investigated whether the S769N mutation influenced the para-
artemether (ATM) to these proteins Since the SmSERCA has
site's sensitivity to ARTs. By using a transfection technique, we
a 263E residue and ATM still kills S. mansoni, it has been argued
replaced the wild-type Pfatp6 allele with the S769N mutant allele
that the residue at 263 may be less critical. Whereas a mutation at
by genetic recombination. Comparison of the resulting parasite
May 2012 Volume 56 Number 5
FIG 2 Pfatp6 expression in wild-type and S769N mutant clones. Pfatp6 ex-
pression levels are shown in the ring (12 h), trophozoite (30 h), and schizont
(38 h) stages. The relative expression level of Pfatp6 was determined by real-
time PCR analysis using primers P3 and P4. A housekeeping gene, seryl-tRNA
synthetase (PF07_0073), was used as an internal control. There were no signif-
icant differences in mRNA levels among the parasite clones at each develop-
ment time point (P ⬎ 0.05, ANOVA).
lines failed to produce significant differences in IC50s to ARTsbetween parasites carrying the wild-type and the mutant alleles,indicating that the S769N mutation of PfATP6 is not involved inmodulating P. falciparum sensitivity to ARTs.
MATERIALS AND METHODS
DNA construct. The transfection construct was designed in the vector
pHD22Y, which carries the human dihydrofolate reductase gene (hdhfr),
conferring resistance to WR99210, with the calmodulin promoter and the
histidine-rich protein 2 terminator A 2.2-kbp fragment upstream of
the stop codon of Pfatp6 was amplified using primers AGATCTCAACAC
CTGTACAATCATCAAATAAG and CTCGAGTTAATCAATTTTAATT
TTCTTGGTTCTTTGC (restriction sites are underlined) and cloned into
a plasmid, Tg23-Luc, at the BglII and XhoI site The fragment con-
FIG 3 Scatter plot of IC s of 3D7 and five transgenic clones carrying either
sisting of the 2,200-bp 3= sequence of the P. berghei dhfr-ts gene (PbDT-3=)
769N (2, 17, and 31) or 769S (9 and 17), assayed with ATM (top), ATS (mid-
was moved into pHD22Y at the BamHI and SpeI site. Site-directed mu-
dle), and DHA (bottom). Each value indicates the mean from three technical
tagenesis was performed to create the S769N mutation by using the
replicates. The means ⫾ standard deviations were calculated from the means
QuikChange Lightning site-directed mutagenesis kit (Agilent Tech-
of 18 biological experiments. For statistical comparison, data were normalized
nologies, La Jolla, CA). Briefly, the plasmid pHD22Y-pfatp6 was am-
using natural logarithm transformation. For each drug, there were no signifi-
plified using two complementary oligonucleotides (5=-GCTTATAAAA
cant differences among the parasite lines after controlling for multiple tests(P ⬎ 0.05, unpaired t test).
5=-CATCTGTATTCTTAATATTTAAATCTTTACTATTTAATTTTTTATAAGC-3=), containing the desired mutation (underlined). After digestionwith KpnI to remove the parental DNA template, the amplified products
parasites were cultured under 5 nM WR99210 for 2 to 3 weeks, until the
were used to transform bacteria, and positive clones were sequenced to
parasitemia reached 5%. This drug on-off cycle was repeated three times,
confirm the presence of the S769N mutation. The plasmid containing the
and resulting parasites were cloned by using the single-cell sorting method
S769N mutation, here designated pHD22Y-pfatp6-769N, was purified for
Confirmation of integration by PCR and Southern blotting. P. fal-
Parasite culture and transfection. The P. falciparum line 3D7, with
ciparum DNA was purified from saponin-released parasites using the phe-
one copy of Pfmdr1, was cultured in human O⫹ erythrocytes at 5%
nol-chloroform extraction method Plasmid integration at the Pfatp6
hematocrit in complete medium (RPMI 1640 supplemented with 25 mM
locus was determined by integration-specific PCR using primer P1 (5=-G
HEPES [pH 7.5], 25 mM sodium bicarbonate, 50 mg/liter hypoxanthine,
ATATATTACCAACATTCTC-3=), located upstream of the integration
0.5% Albumax II, and 40 g/ml gentamicin sulfate) as previously de-
region, and P2 (5=-CATATCCGGTACCATTGTC-3=) located in PbDT-3=
scribed Cultures were maintained at 37°C in a gas mixture of 5%
of the plasmid PCR products from different parasites were
CO , 3% O , and 92% N . Culture synchronization was performed by two
sequenced to identify clones with the S769N mutation To con-
rounds of treatment of ring-stage parasites with 5% (vol/vol) sorbitol for
firm the integration event at the Pfatp6 locus, Southern hybridization was
5 min Parasites were released by treatment with 0.05% saponin.
performed as previously described Briefly, 3 g of parasite DNA
Transfection of the parasite was performed using the erythrocyte loading
from each clone was digested with KpnI, separated on a 1% agarose gel,
method After transfection, parasites were cultured under 2.5 nM
and transferred to a nylon membrane. The probe located upstream of the
WR99210 until resistant parasites emerged and reached 5% parasitemia.
integration region was amplified with primers (5=-GCTGCCGT
To enrich parasites with chromosomal integration of the plasmid, para-
AGGTGTATG-3= and 5=-CCATGAATTGGATCTGAG-3=) and labeled
sites were cultured in the absence of drug for 2 weeks. Afterwards, the
with digoxigenin (DIG) by using a DIG PCR labeling kit (Roche Applied
Antimicrobial Agents and Chemotherapy
PfATP6 and Artemisinin Resistance in P. falciparum
TABLE 1 Unpaired t test results for mutant vs control lines exposed to
tion is able to modulate susceptibility of P. falciparum to ARTs, we
generated transgenic parasites lines expressing the S769N mutant
P valuea
PfATP6 in 3D7 by using a single-crossover strategy Aftertransfection of the 3D7 parasite with the pHD22Y-pfatp6-769N
769N-2 vs 769S-17
construct, parasites were selected with WR99210 until resistant
parasites appeared in 3 weeks. Afterwards, parasites were culturedthrough three drug on-off cycles to enrich parasites with chromo-
769N-31 vs 769S-17
somal integration of the plasmid. To obtain multiple mutant
clones with the S769N mutation, 200 clones were obtained by
single cell sorting and analyzed. Integration-specific PCR usingprimers P1 and P2 identified 32 positive clones with the
correct integration event occurring at the Pfatp6 locus
After sequencing each PCR product of the 32 clones, three clones
(769N-2, 769N-7, and 769N-31) were found to harbor the S769N
After a Bonferonni correction, P values of less than 0.0056 were considered significant.
mutation Since genetic recombination with single cross-over often results in the insertion of plasmid concatemers, we
Science). The membrane was hybridized with denatured probes for 12 h at58°C. Hybridized DNA was detected with a DIG luminescence detectionkit (Roche Applied Science) and exposed to X-ray film.
To further determine whether the single-crossover events involved the
integration of plasmid concatemers, the plasmid copy numbers in theintegrated clones were determined by real-time PCR analysis using prim-ers P3 (5=-GTTTTCTGTAGAACTG-3=) and P4 (5=-GATAACGGATAAATGC-3=) Pfapt6 copy number was determined by comparingit with 3D7 using the 2⌬⌬CT method with the single-copy gene seryl-tRNAsynthetase (PF07_0073) as an internal reference
Pfatp6 gene expression. To assess the expression level of Pfatp6 in
different parasite clones, total RNA was isolated from synchronized par-asites at the ring (12 h), trophozoite (30 h), and schizont (38 h) stages byusing TRIzol (Invitrogen, Carlsbad, CA). The RNA was directly used astemplate for real-time reverse transcriptase PCR (RT-PCR) analysis usinga One-Step quantitative RT-PCR master mix kit (USB, Cleveland, OH).
The relative expression level was calculated by comparing the result withthat in 3D7 and using the 2⌬⌬CT method The housekeeping geneseryl-tRNA synthetase (PF07_0073) was used as an internal reference.
In vitro drug assays. Three ART derivatives, DHA, ATM, and ATS,
were purchased from Sigma (St. Louis, MO). Drug stock solutions (10mM) were made fresh in dimethyl sulfoxide (DMSO) and stored at
⫺80°C. In vitro sensitivities of the parasite lines to ART derivatives weredetermined by using the SYBR green I method with 2-fold serial dilutionsof the drugs to final concentrations of 0.8 to 200 nM Briefly, 100
l of a ring-stage parasite culture at 0.5% parasitemia and 1% hematocritin the culture medium with different drug concentrations was seeded intriplicate in 96-well flat-bottom plates and incubated at 37°C for 72 h.
Afterwards, the plates were frozen and thawed, mixed with 100 l of lysisbuffer, and incubated in the dark at 37°C for 4 h. Fluorescence was mea-sured using the FLUOstar OPTIMA microplate reader (BMA Labtech,Offenburg, Germany) with excitation and emission wavelengths centeredat 485 and 538 nm, respectively. For accuracy, each parasite line was mea-sured in 18 biological replicates, each with three technical replicates. IC s
were calculated using the program GraphPad Prism version 5 (La Jolla,CA) by constructing a dose-response curve. The percentage of inhibitionwas calculated using the following formula: [(fluorescence of drug treatedparasites ⫺ fluorescence of untreated control)/(fluorescence of untreatedcontrol)] ⫻100.
Statistical analysis. The mean IC s of mutant parasite clones were
compared to the mean IC s of control lines by using unpaired t tests,
assuming unequal variances. The data were first transformed using thenatural logarithm in order to control for nonnormality in IC s. In order
to control for multiple tests, both a Bonferroni correction and a Benja-mini-Hochberg correction were applied to the t test results.
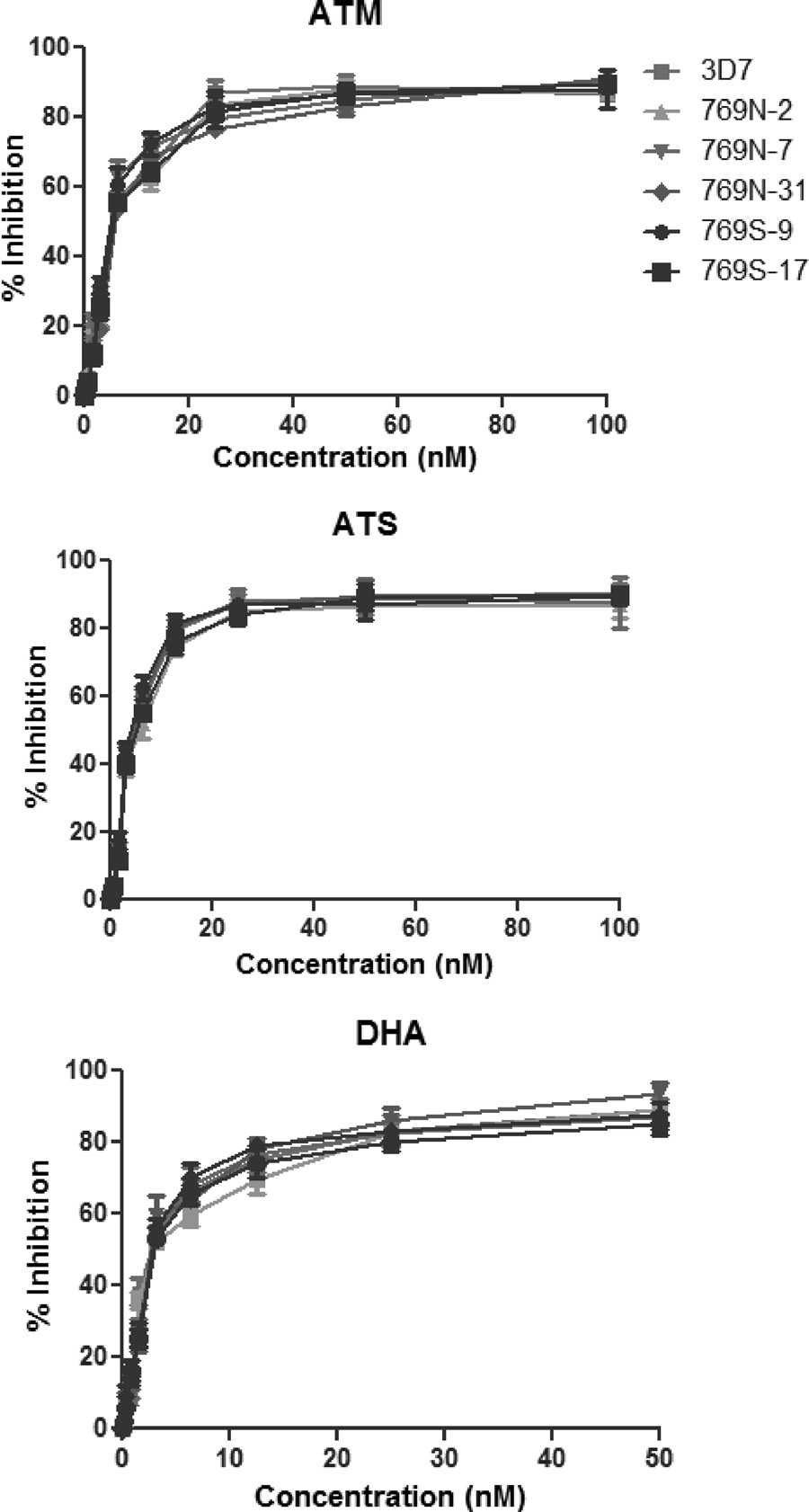
FIG 4 Dose-response curves of 3D7 and five parasite clones with either 769N
(2, 17, and 31) or 769S (9 and 17) assayed against ATM (top), ATS (middle),and DHA (bottom). The results were obtained from 18 independent experi-
Development of transgenic lines expressing the PfATP6 S769N
ments, each with three technical replicates. Percent inhibition values are
mutation. To determine whether PfATP6 with the S769N muta-
shown as means ⫾ standard deviations.
May 2012 Volume 56 Number 5
determined the copy number of the integrated plasmid in all 34
ARTs in the Xenopus oocyte system The L263E mutation has
clones by real-time PCR analysis. The result showed that clones
so far eluded detection in field parasite populations, and introduc-
769N-2 and 769N-31 had one copy of the plasmid inserted into
tion of L263E in P. falciparum through allele exchange did not
the genome, whereas 769N-7 had three copies of the plasmid. To
cause significant changes of parasite sensitivities to ARTs
eliminate the possible effects of insertion of different copies of the
While such a discrepancy is not clearly understood, a recent dock-
plasmid on sensitivities to ARTs, two parasite clones (769S-17 and
ing simulation study suggested that the significance of L263E in
769S-9) from similar integration events but without the S769N
ART resistance may be less than previously hypothesized The
mutation were chosen as transfection controls. Clones 769S-17
S769N mutation was originally found in P. falciparum field iso-
and 769S-9 had one and three copies of the plasmid integrated,
lates from French Guiana, and it was linked to increased resistance
respectively The integration events of the selected five
to ATM Although the S769N mutation has been found in
clones were further confirmed by Southern blotting As
some parasite isolates from Africa a drug assay on a
predicted, a 10-kb KpnI fragment was detected in 3D7 parasite
single parasite isolate showed that it was sensitive to DHA and
genomic DNA, whereas a 5.8-kb KpnI fragment was observed
ATM In this study, we showed that introduction of the
after the integration of the plasmid at the Pfatp6 locus
S769N mutation in 3D7 by an allele exchange strategy did not alter
Pfatp6 gene expression. To determine whether the integration
the parasite's sensitivity to all tested ART derivatives. Whereas
events affected Pfatp6 expression, the mRNA levels of Pfatp6 in the
these data argue against the predicted role of these PfATP6 muta-
five selected clones were compared using real-time RT-PCR with
tions in modulating ART sensitivity, it remains to be determined
3D7 as the reference. No significant difference in Pfatp6 expres-
whether the divergent findings are due to different genetic back-
sion was detected among the five clones (P ⬎ 0.05, ANOVA)
grounds of the parasite lines. It has been reported that genetic
This result was consistent with the prediction, since for
backgrounds of the parasites may greatly influence the effect of
clones with insertion of more than one copy of the plasmid, only
Pfcrt on CQ resistance Pfmdr1 on resistance to CQ and qui-
the first copy of the Pfatp6 gene had a promoter and was tran-
nine and Pfnhe1 on quinine resistance
Sequencing of PfATP6 has identified many mutations in this
In vitro response of transgenic lines to ART derivatives. We
gene among field parasite populations but none of them have
next determined the IC50s of 3D7, three mutant lines (769N-2, -7,
been conclusively linked to ART resistance. Some studies showed
and -31) and two control lines (769S-9 and -17) to the three ART
that deployments of ACTs were associated with changes of fre-
derivatives. The IC50 of each parasite clone to each of the ART
quencies of certain mutations in PfATP6. In one study the fre-
derivatives was determined in 18 biological replicates, each with
quency of the A623E mutation was increased in Niger after ACT
three technical replications All transfectant lines and 3D7
use whereas in another study an increase in the frequency of a
had similar IC50s against ATM, ATS, and DHA. The means of
deletion mutant was noticed in Peru In the GMS, an
absolute IC50s of the five transgenic lines were log transformed
epicenter of malaria drug resistance with the most extensive use of
and compared for statistical significance. Statistical analysis con-
ART drugs, A623E and S769N mutations associated with reduced
firmed the lack of a significant difference in IC50s between mutant
sensitivity to ARTs have not been detected so far In addition,
lines and their control lines (P ⬎ 0.0056, unpaired t
the clinical ART resistance in western Cambodia is not associated
tests). We further compared the dose-response curves of all para-
with PfATP6 It is noteworthy that most of the PfATP6 mu-
site lines and found that the dose-response patterns for the ART
tations are rare and geographically confined. Molecular evolution
derivatives were very similar
analysis of PfATP6 single-nucleotide polymorphisms showed thatthe ratio of synonymous versus nonsynonymous substitutions did
not significantly deviate from neutrality Moreover, our
The mode of action of ARTs in malaria parasites is still not com-
analysis of parasite samples collected from the GMS after deploy-
pletely understood, and the molecular basis of reduced ART sus-
ment of ARTs revealed similar findings Collectively, the ev-
ceptibility is unclear So far, a number of genes have
idence accumulated thus far strongly suggests that PfATP6 does
been proposed to be associated with reduced sensitivities to ARTs,
not have much to do with ART resistance.
but none of the associations has been conclusively validated Based on heterologous expression studies and biochemical assays,
PfATP6 has been postulated to be a prime target of ART, and
This work was supported by NIAID, NIH (1R21AI085518 and
L263E was considered a potential mutation that mediatesART re-
U19AI089672) and by the Intramural Research Program of the Division
sistance These initial studies spurred extensive investiga-
of Intramural Research, National Institute of Allergy and Infectious Dis-
tions on PfATP6 but results obtained so far have cast
eases, National Institutes of Health.
considerable doubts on the role of PfATP6 in ART resistance.
Biochemical studies of purified PfATP6 failed to detect inhibition
of this enzyme by ARTs suggesting that findings from het-
1. Anderson TJ, et al. 2005. Are transporter genes other than the chloro-
erologous expression in Xenopus oocytes may be a system-specific
quine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) as-sociated with antimalarial drug resistance? Antimicrob. Agents Che-
effect. In this study, we further evaluated the potential role of
mother. 49:2180 –2188.
PfATP6 in ART resistance, and our allele exchange experiments
2. Arnou B, et al. 2011. The Plasmodium falciparum Ca(2⫹)-ATPase
confirmed the lack of association of the S769N mutation of this
PfATP6: insensitive to artemisinin, but a potential drug target. Biochem.
gene with altered sensitivity to ARTs.
Soc. Trans. 39:823– 831.
3. Bacon DJ, et al. 2009. Dynamics of malaria drug resistance patterns in the
Two mutations in PfATP6 at positions 263 and 769 have been
Amazon basin region following changes in Peruvian national treatment
linked to ART resistance. The L263E mutation was proposed
policy for uncomplicated malaria. Antimicrob. Agents Chemother. 53:
based on docking simulation and found to confer insensitivity to
Antimicrobial Agents and Chemotherapy
PfATP6 and Artemisinin Resistance in P. falciparum
4. Bhisutthibhan J, et al. 1998. The Plasmodium falciparum translationally
Plasmodium falciparum Ca2⫹-ATPase (PfATP6) and docking of artemis-
controlled tumor protein homolog and its reaction with the antimalarial
inin derivatives to PfATP6. Bioorg. Med. Chem. Lett. 15:2994 –2997.
drug artemisinin. J. Biol. Chem. 273:16192–16198.
30. Klonis N, et al. 2011. Artemisinin activity against Plasmodium falciparum
5. Bosman A, Mendis KN. 2007. A major transition in malaria treatment:
requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. U. S. A.
the adoption and deployment of artemisinin-based combination thera-
pies. Am. J. Trop. Med. Hyg. 77:193–197.
31. Krishna S, Pulcini S, Fatih F, Staines H. 2010. Artemisinins and the
6. Briolant S, et al. 2011. In vitro susceptibility to quinine and microsatellite
biological basis for the PfATP6/SERCA hypothesis. Trends Parasitol. 26:
variations of the Plasmodium falciparum Na⫹/H⫹ exchanger (Pfnhe-1)
gene: the absence of association in clinical isolates from the Republic of
32. Krishna S, Uhlemann AC, Haynes RK. 2004. Artemisinins: mechanisms
Congo. Malar. J. 10:37.
of action and potential for resistance. Drug Resist. Update 7:233–244.
7. Cardi D, et al. 2010. Purified E255L mutant SERCA1a and purified
33. Krishna S, Woodrow CJ, Staines HM, Haynes RK, Mercereau-Puijalon
PfATP6 are sensitive to SERCA-type inhibitors but insensitive to artemis-
O. 2006. Re-evaluation of how artemisinins work in light of emerging
inins. J. Biol. Chem. 285:26406 –26416.
evidence of in vitro resistance. Trends Mol. Med. 12:200 –205.
8. Chaijaroenkul W, Pruktal P, Muhamad P, Na-Bangchang K. 2007. Assess-
34. Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium fal-
ment of in vitro antimalarial interactions between dihydroartemisinin and
ciparum erythrocytic stages in culture. J. Parasitol. 65:418 – 420.
fosmidomycin. Southeast Asian J. Trop. Med. Public Health 38:791–795.
35. Lepore R, et al. 2011. Identification of the Schistosoma mansoni molec-
9. Charoenteeraboon J, Kamchonwongpaisan S, Wilairat P, Vattanavi-
ular target for the antimalarial drug artemether. J. Chem. Infect. Model.
boon P, Yuthavong Y. 2000. Inactivation of artemisinin by thalassemic
erythrocytes. Biochem. Pharmacol. 59:1337–1344.
36. Li W, et al. 2005. Yeast model uncovers dual roles of mitochondria in
10. Chavchich M, et al. 2010. Role of pfmdr1 amplification and expression in
action of artemisinin. PLoS Genet. 1:e36.
induction of resistance to artemisinin derivatives in Plasmodium falcipa-
37. Meng H, et al. 2010. In vitro sensitivity of Plasmodium falciparum clinical
rum. Antimicrob. Agents Chemother. 54:2455–2464.
isolates from the China-Myanmar border area to quinine and association
11. Cojean S, Hubert V, Le Bras J, Durand R. 2006. Resistance to dihydro-
with polymorphism in the Na⫹/H⫹ exchanger. Antimicrob. Agents Che-
artemisinin. Emerg. Infect. Dis. 12:1798 –1799.
mother. 54:4306 – 4313.
12. Cojean S, Hubert V, Le Bras J, Durand R. 2007. Resistance to dihydro-
38. Meshnick SR. 2002. Artemisinin: mechanisms of action, resistance and
artemisinin: in response. Emerg. Infect. Dis. 13:809.
toxicity. Int. J. Parasitol. 32:1655–1660.
13. Cui L, Fan Q, Li J. 2002. The malaria parasite Plasmodium falciparum
39. Meshnick SR. 1994. The mode of action of antimalarial endoperoxides.
encodes members of the Puf RNA-binding protein family with conserved
Trans. R. Soc. Trop. Med. Hyg. 88(Suppl. 1):S31–S32.
RNA binding activity. Nucleic Acids Res. 30:4607– 4617.
40. Meshnick SR, et al. 1993. Iron-dependent free radical generation from
14. Cui L, Miao J, Cui L. 2007. Cytotoxic effect of curcumin on malaria
the antimalarial agent artemisinin (qinghaosu). Antimicrob. Agents Che-
parasite Plasmodium falciparum: inhibition of histone acetylation and
mother. 37:1108 –1114.
generation of reactive oxygen species. Antimicrob. Agents Chemother.
41. Miao J, Cui L. 2011. Rapid isolation of single malaria parasite-infected red
51:488 – 494.
blood cells by cell sorting. Nat. Protoc. 6:140 –146.
15. Cui L, Miao J, Wang J, Li Q, Cui L. 2008. Plasmodium falciparum:
42. Miao J, Li J, Fan Q, Li X, Cui L. 2010. The Puf-family RNA-binding
development of a transgenic line for screening antimalarials using firefly
protein PfPuf2 regulates sexual development and sex differentiation in the
luciferase as the reporter. Exp. Parasitol. 120:80 – 87.
malaria parasite Plasmodium falciparum. J. Cell Sci. 123:1039 –1049.
16. Cui L, Su XZ. 2009. Discovery, mechanisms of action and combination
43. Reference deleted.
therapy of artemisinin. Expert Rev. Anti Infect. Ther. 7:999 –1013.
44. Naik PK, et al. 2011. The binding modes and binding affinities of arte-
17. Dahlstrom S, et al. 2008. Diversity of the sarco/endoplasmic reticulum
misinin derivatives with Plasmodium falciparum Ca2⫹-ATPase (PfATP6).
Ca(2⫹)-ATPase orthologue of Plasmodium falciparum (PfATP6). Infect.
J. Mol. Model. 17:333–357.
Genet. Evol. 8:340 –345.
45. Noedl H, et al. 2008. Evidence of artemisinin-resistant malaria in western
18. Deitsch K, Driskill C, Wellems T. 2001. Transformation of malaria
Cambodia. N. Engl. J. Med. 359:2619 –2620.
parasites by the spontaneous uptake and expression of DNA from human
46. Nosten F. 2010. Waking the sleeping beauty. J. Infect. Dis. 202:1300 –1301.
erythrocytes. Nucleic Acids Res. 29:850 – 853.
47. Nosten F, White NJ. 2007. Artemisinin-based combination treatment of
19. Ding XC, Beck HP, Raso G. 2011. Plasmodium sensitivity to artemis-
falciparum malaria. Am. J. Trop. Med. Hyg. 77:181–192.
inins: magic bullets hit elusive targets. Trends Parasitol. 27:73– 81.
48. O'Neill PM, Barton VE, Ward SA. 2010. The molecular mechanism of
20. Dondorp AM, et al. 2009. Artemisinin resistance in Plasmodium falcipa-
action of artemisinin: the debate continues. Molecules 15:1705–1721.
rum malaria. N. Engl. J. Med. 361:455– 467.
49. Phompradit P, Wisedpanichkij R, Muhamad P, Chaijaroenkul W, Na-
21. Eckstein-Ludwig U, et al. 2003. Artemisinins target the SERCA of Plas-
Bangchang K. 2011. Molecular analysis of pfatp6 and pfmdr1 polymor-
modium falciparum. Nature 424:957–961.
phisms and their association with in vitro sensitivity in Plasmodium falcipa-
22. Eshetu T, et al. 2010. Different mutation patterns of Plasmodium falcip-
rum isolates from the Thai-Myanmar border. Acta Trop. 120:130 –135.
arum among patients in Jimma University Hospital, Ethiopia. Malar. J.
50. Reed MB, et al. 2000. Targeted disruption of an erythrocyte binding
antigen in Plasmodium falciparum is associated with a switch toward a
23. Fidock DA, Wellems TE. 1997. Transformation with human dihydrofo-
sialic acid-independent pathway of invasion. Proc. Natl. Acad. Sci. U. S. A.
late reductase renders malaria parasites insensitive to WR99210 but does
not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U. S. A.
51. Roper C, et al. 2004. Intercontinental spread of pyrimethamine-resistant
malaria. Science 305:1124.
24. Hunt P, et al. 2007. Gene encoding a deubiquitinating enzyme is mutated
52. Shahinas D, Lau R, Khairnar K, Hancock D, Pillai DR. 2010. Artesunate
in artesunate- and chloroquine-resistant rodent malaria parasites. Mol.
misuse and Plasmodium falciparum malaria in traveler returning from
Microbiol. 65:27– 40.
Africa. Emerg. Infect. Dis. 16:1608 –1610.
25. Ibrahim ML, Khim N, Adam HH, Ariey F, Duchemin JB. 2009. Poly-
53. Sidhu AB, Valderramos SG, Fidock DA. 2005. pfmdr1 mutations con-
morphism of PfATPase in Niger: detection of three new point mutations.
tribute to quinine resistance and enhance mefloquine and artemisinin
Malar J. 8:28.
sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913–926.
26. Imwong M, et al. 2010. Exploring the contribution of candidate genes to
54. Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004.
artemisinin resistance in Plasmodium falciparum. Antimicrob. Agents
Simple and inexpensive fluorescence-based technique for high-
Chemother. 54:2886 –2892.
throughput antimalarial drug screening. Antimicrob. Agents Chemother.
27. Jambou R, et al. 2005. Resistance of Plasmodium falciparum field isolates
to in-vitro artemether and point mutations of the SERCA-type PfAT-
55. Tanabe K, et al. 2011. Spontaneous mutations in the Plasmodium falciparum
Pase6. Lancet 366:1960 –1963.
sarcoplasmic/endoplasmic reticulum Ca2⫹-ATPase (PfATP6) gene among
28. Jambou R, et al. 2010. Geographic structuring of the Plasmodium falcip-
geographically widespread parasite populations unexposed to artemisinin-
arum sarco(endo)plasmic reticulum Ca2⫹ ATPase (PfSERCA) gene di-
based combination therapies. Antimicrob. Agents Chemother. 55:94 –100.
versity. PLoS One 5:e9424.
56. Uhlemann AC, et al. 2005. A single amino acid residue can determine the
29. Jung M, Kim H, Nam KY, No KT. 2005. Three-dimensional structure of
sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 12:628 – 629.
May 2012 Volume 56 Number 5
57. Valderramos SG, et al. 2010. Identification of a mutant PfCRT-mediated
60. White NJ. 2008. Qinghaosu (artemisinin): the price of success. Science
chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pat-
61. WHO. 2011. World malaria report 2010. World Health Organization,
58. Vattanaviboon P, Wilairat P, Yuthavong Y. 1998. Binding of dihydro-
artemisinin to hemoglobin H: role in drug accumulation and host-
induced antimalarial ineffectiveness of alpha-thalassemic erythrocytes.
62. Witkowski B, et al. 2010. Increased tolerance to artemisinin in Plasmo-
Mol. Pharmacol. 53:492– 496.
dium falciparum is mediated by a quiescence mechanism. Antimicrob.
59. Wang J, et al. 2010. Artemisinin directly targets malarial mitochondria
Agents Chemother. 54:1872–1877.
through its specific mitochondrial activation. PLoS One 5:e9582.
63. Wootton JC, et al. 2002. Genetic diversity and chloroquine selective
59a.Wang Z, et al. 2010. In vitro sensitivity of Plasmodium falciparum from
sweeps in Plasmodium falciparum. Nature 418:320 –323.
China-Myanmar border area to major ACT drugs and polymorphisms in
64. Zhang G, Guan Y, Zheng B, Wu S, Tang L. 2008. No PfATPase6 S769N
potential target genes. PLoS One, in press.
mutation found in Plasmodium falciparum isolates from China. Malar. J. 7:122.
Antimicrobial Agents and Chemotherapy
Source: http://www-personal.ksu.edu/~hwang/research/10.Cui.Wang.Jiang.Parker.Wang.Su.Cui.Lack.of.Association.2012.pdf
"Farmacología kinésica deportiva" Cátedra Kinesiología Deportiva Encargado de enseñanza Dr. Mastrángelo, Jorge Lic. Spinetta, Daniel Integrantes Balzi, Brenda Bettini, Florencia Ferraris, Juan Manuel Fortuondo, María Emilce Gómez, Vanina Guisasola, Pablo L'Afflitto, Mariana Micó, Gustavo Vazquez, Lorena Vignolo, Florencia
Comp. Biochem. Physiol. Vol. 116B, No. 2, pp. 269–277, 1997 ISSN 0305-0491/ 97/$17.00 Copyright 1997 Elsevier Science Inc. Seasonal Levels of Reproductive Hormones and Their Relationship to the Antler Cycle of Male and Female Reindeer (Rangifer tarandus) George A. Bubenik,a Dieter Schams,b Robert J. White,c Janice Rowell,c John Blake,d and Ludek Bartosd



