Ufv.br
Proc. Natl. Acad. Sci. USA
Vol. 95, pp. 9009–9013, July 1998
Ferredoxin-1 mRNA is destabilized by changes in photosynthetic
electron transport
MARIE E. PETRACEK*†, LYNN F. DICKEY*, TUYEN T. NGUYEN‡, CHRISTIANE GATZ§, DOLORES A. SOWINSKI*,
GEORGE C. ALLEN*, AND WILLIAM F. THOMPSON*Departments of *Botany and ‡Crop Science, North Carolina State University, Raleigh, NC 27695; and §Albrecht von haller Institute for Plant Sciences, University
of Gottingen, Untere Karspule 2, 37073 Gottingen, Germany
Edited by Elisabeth Gantt, University of Maryland at College Park, College Park, MD, and approved May 29, 1998 (received for review
March 12, 1998)
In transgenic tobacco, pea Ferredoxin-1
tional initiation context also compromises light regulation of
(Fed-1) mRNA accumulates rapidly in response to photosyn-
the
Fed-1 mRNA (7). Finally, the
Fed-1 mRNA is polyribo-
thesis even when the transgene is driven by a constitutive
some-associated in the light but not in the dark (8), thus
promoter. To investigate the role of photosynthesis on Fed-1
decreased
Fed-1 mRNA abundance is correlated with a de-
mRNA stability, we used the tetracycline repressible Top10
crease in
Fed-1 translation rates.
promoter system to specifically shut off transcription of the
Fed-1 mRNA levels also decrease when light-grown plants
Fed-1 transgene. The Fed-1 mRNA has a half-life of approx-
are treated with the photosynthetic electron transport inhibitor
imately 2.4 hr in the light and a half-life of only 1.2 hr in the
dark or in the presence of the photosynthetic electron trans-
reillumination of DCMU-treated plants after a 40-hr dark
treatment,
Fed-1 mRNA fails to reaccumulate and remains
(DCMU). These data indicate that cessation of photosynthe-
dissociated from polyribosomes (8). These results suggest light
sis, either by darkness or DCMU results in a destabilization
regulation of
Fed-1 mRNA accumulation and translation
of the Fed-1 mRNA. Furthermore, the Fed-1 mRNA half-life is
requires photosynthetic electron transport.
reduced immediately upon transfer to darkness, suggesting
It is of interest to know whether the
Fed-1 mRNA half-life
that Fed-1 mRNA destabilization is a primary response to
is altered by changes in photosynthetic activity. One method of
photosynthesis rather than a secondary response to long-term
mRNA half-life determination is to globally inhibit transcrip-
dark adaptation. Finally, the two different methods for effi-
tion and monitor the decay of the mRNA of interest over time.
cient tetracycline delivery reported here generally should be
This approach has been used successfully in plants to give
useful for half-life measurements of other mRNAs in whole
half-lives that correspond to changes in mRNA steady-state
abundance (9, 10). However, apparent secondary effects from
global transcriptional inhibition confounded half-life studies in
The pea Ferredoxin-1 (
Fed-1) gene encodes plastid-localized
other cases (11, 12).
photosystem I ferredoxin. Its mRNA is perhaps the best-
Attempts to use actinomycin D (ActD) to determine
characterized nuclear-encoded transcript in plants that exhib-
whether
Fed-1 mRNA half-life is altered in the light have been
its promoter-independent regulation of mRNA abundance
unsuccessful (13). In these experiments, ActD treatment re-
(e.g., refs. 1 and 2).
Fed-1 initially was identified in a cDNA
sulted in an increase rather than the expected decrease in
screen for light-regulated mRNAs in pea (3) and was chosen
Fed-1 mRNA in the dark, preventing a determination of
Fed-1
for further study because
Fed-1 mRNA accumulated faster
mRNA half-life. There are at least two ways in which these
than several other light-regulated mRNAs in etiolated pea
results can be reconciled with our observation that the steady-
seedlings exposed to light (4). Analysis of transgenic tobacco
state abundance of
Fed-1 mRNA declines in darkness in a
plants containing the transcribed sequence of the pea
Fed-1
promoter-independent manner. First, the
Fed-1 mRNA may
gene under the transcriptional control of the constitutive
indeed be more stable in darkness than in light, but this effect
cauliflower mosaic virus 35S promoter (P
could be overridden if transcriptional elongation is inhibited in
35S) revealed that the
light-regulated accumulation of
Fed-1 mRNA was conferred
the dark. This transcriptional effect on abundance may not be
by the mRNA sequence itself rather than by the promoter (1).
detectable by transcriptional run-on assays (reviewed in ref.
Furthermore, the light effect on accumulation of
Fed-1 mRNA
14). Second, global inhibition of transcription might lead to
transcribed from this chimeric gene was greater in green plants
improper
Fed-1 light regulation—for example, by causing the
than in etiolated seedlings exposed to light for the first time
disappearance of a labile factor required for
Fed-1 degradation
(5). Deletion analysis determined that 95 nt of the 59 UTR plus
in the dark.
the first one-third of the
Fed-1 coding region ('143 nt) are
The secondary effects caused by global transcriptional in-
sufficient for light regulation in such constructs. This region is
hibitors can be circumvented by making mRNA half-life
referred to as the internal light regulatory element (iLRE) (6).
measurements under conditions in which transcription is sup-
Further deletion of the
Fed-1 iLRE results in a gradual loss of
pressed only for the gene of interest, while all other genes are
light regulation (7).
transcribed normally. The tetracycline (tet)yVP16 system used
Translation appears to be involved in the
Fed-1 mRNA light
in animals has been adapted for use in plants (15). The Top10
response. Mutation of the
Fed-1 initiation codon to either a
tetracycline-repressible promoter (P-Top10) is based on the
missense or nonsense codon abolishes light regulation of the
constitutive P35S, but contains seven tetracycline operator sites
mRNA (2). Similarly, mutation of the surrounding transla-
in place of the upstream activator sequences. Plants are
The publication costs of this article were defrayed in part by page charge
This paper was submitted directly (Track II) to the
Proceedings office.
payment. This article must therefore be hereby marked ‘‘
advertisement'' in
Abbreviations: Fed-1, Ferredoxin-1; DCMU, 3-(3,4-dichlorophenyl)-
1,1-dimethylurea; P
accordance with 18 U.S.C. §1734 solely to indicate this fact.
35S, caulif lower mosaic virus 35S promoter; ActD,
actinomycin D.
1998 by The National Academy of Sciences 0027-8424y98y959009-5$2.00y0
†To whom reprint requests should be addressed. e-mail:
PNAS is available online at http:yywww.pnas.org.
Plant Biology: Petracek
et al.
Proc. Natl. Acad. Sci. USA 95 (1998)
transformed with a construct encoding the tet repressor fused
liquid on top of the nylon membrane was removed and
to the VP16 transcriptional activator protein and with a
replaced with 3 ml of 10 mgyliter tetracycline in Hoagland's
construct containing the P-Top10 fused to the
GUS reporter
solution with or without 1 mM DCMU. Time zero was
gene. In the absence of tetracycline, the tet repressor binds the
designated as the time after 1 hr of tetracycline solution
P-Top10 and VP16 activates
GUS transcription. Upon addition
uptake; at this time, covers were replaced and boxes were
of tetracycline, the tetyVP16 fusion protein no longer binds
either wrapped in foil for dark time points, or left in the light
P-Top10, preventing transcription of
GUS (16). Thus, this
for light or DCMUylight time points. At appropriate time
system can be used for a direct measurement of the half-life of
points, leaves from all 15–20 plants grown in a single box were
the
GUS mRNA or any other mRNA driven by P-Top10. The
harvested into liquid nitrogen.
Top10 system has been used successfully to measure half-life
Octuplet Production. Plantlets derived from transgenic calli
of SAUR containing mRNAs in tobacco suspension culture
were successively subdivided to produce eight clonal plantlets
(octuplets) expressing
Fed-1. Octuplets were rooted on rooting
We have observed that a normal
Fed-1 mRNA light re-
medium, then transferred to Plant Cons (Sigma) containing
sponse requires healthy, intact green plants. Previously, in
approximately 20 ml of Hoagland's solution and placed in a
studies of the half-life of GUS mRNA using the P-Top10
growth chamber illuminated at an intensity of 240 mmol
system, tetracycline was introduced into detached leaves by
m22zs21 supplied by a mixture of 6 incandescent and 12
vacuum infiltration (16). On the whole-plant level, the half-life
fluorescent lights on a 12-hr lighty12-hr dark cycle. Plants were
of Gus protein, not mRNA, was examined over a period of
supplied with fresh Hoagland's solution every other day for a
days, not hours, using low levels of tetracycline. To study the
week to allow plants to grow and to adapt to the new light
rapid
Fed-1 mRNA light response, it was essential that we
conditions. Half-life measurements were carried out as above
develop conditions permitting rapid delivery of tetracycline to
except that the entire 20 ml of Hoagland's solution in the Plant
intact, healthy plants. Here we report measuring
Fed-1 mRNA
Cons was completely replaced with 20 ml of fresh Hoagland's
half-life from intact plants by using P-Top10. We have deter-
solution supplemented with 10 mgyliter tet andyor 1 mM
mined that the half-life of the
Fed-1 mRNA is significantly
DCMU. A single member of the octuplet set was harvested for
shortened in the dark and in the presence of photosynthetic
each time point.
electron transport inhibitor DCMU. These results show that
Polyribosome Profiles. Plants were grown on nylon mem-
light regulates
Fed-1 mRNA abundance by affecting its sta-
branes as above, treated with the solutions and light regime
indicated, and harvested into liquid nitrogen. Polyribosomes
then were isolated on a 15–65% sucrose gradient as previously
described, and the absorbance of the fractions at UV 254 nm
was monitored and recorded during fractionation.
Gene Construct. The P-Top10::
Fed-1 construct was derived
from the ‘‘message'' construct as described in ref. 1. An
XbaI
and
EcoRI (blunt-ended with Klenow) fragment, which in-
cluded the Fed-1 transcribed region (749 bp) and 70 bp of the
To make the P-Top10 useful for the examination of
Fed-1
39 flanking region, was substituted for the GUS-INTyoctopine
half-life, we needed to develop a system for rapid and efficient
synthase terminator fragment [in pTOP10 (16)], which was
excised by using
XbaIy
HindIII (blunt-ended with Klenow).
uptake of tetracycline in healthy plants. Thus, we developed
The resulting plasmid was transferred to
Agrobacterium tume-
the P-Top10 repressible system for use in two different types
faciens strain LBA4404 by triparental mating (18).
of plant growth systems as follows: Tobacco Petite havana was
transformed sequentially with DNA encoding the VP16
Plant Transformations. Nicotiana tabacum (SR-1, Petite
Havana) were transformed by using the leaf disc method (19).
activator and then with the P-Top10::
Fed-1::
Fed-1 ter con-
Initially, plants were transformed with the transactivator plas-
struct (Fig. 1). Transgenic plant lines were screened for
mid pTetVP16 (16) and selected on kanamycin (300 mgyml).
expression of the
Fed-1 mRNA, which indicates appropriate
The transformant expressing the highest amount of TetVP16
expression of both transgenes because the VP16yTet gene
mRNA was chosen for a second round of transformation with
product is required for activation of the P-Top10. Most lines
the P-Top10::
Fed-1 construct described above, and transfor-
had significant levels of expression. One line (5010) was chosen
mants were selected on hygromycin (50 mgyml).
for further study, and T2 seeds were collected for the exper-
Growth of Transgenic Tobacco on Nylon Membrane. Twen-
iments with seedlings grown hydroponically on nylon mem-
ty-five milliliters of sterile MS medium was added to thor-
branes. Fifteen other transgenic plant lines produced from a
oughly rinsed and autoclaved magenta boxes with membrane
raft units (Sigma V8380; Sigma M1917). T2 transgenic tobacco
seeds (line 5010) were sterilized in 30% commercial bleach
followed by five washes with sterile deionized water. Approx-
imately 15–20 transgenic seeds were spread on each mem-
brane, the magenta box lids were put back on the box, and the
junction between the lid and box was wrapped with micropore
tape (3M Co., 1530–0). Plants were grown sterilely for 3–4
weeks to at least a four-leaf stage in a 20°C growth chamber.
White light (120 mmol m22zs21) was supplied by a mixture of
six incandescent and six fluorescent lights operating on a 12-hr
lighty12-hr dark cycle.
Half-Life Measurements in Seedlings. Two hours after the
FIG. 1. Constructs used in the repressible promoter system. The
beginning of the light cycle, transgenic tobacco plants were
chimeric transactivator Tet-VP16 is expressed under control of the
treated either with 3 ml of 1 mM DCMU in Hoagland's
cauliflower mosaic virus 35S promoter (P35S) (
Upper). Progeny of
solutiony1% EtOH or with Hoagland's solutiony1% EtOH
plants transformed with the transactivator construct have been re-
transformed with Fed-1 under control of the synthetic Top10 promoter
(control plants). These solutions were added to roots growing
(P-Top10) (
Lower). The Tet-VP16 protein binds P-Top10, allowing
across the nylon membrane, and the Magenta Box covers
transcription to
Fed-1. Endogenously supplied tetracycline binds Tet-
propped open on the top to permit air exchange and encourage
VP16, making Tet-VP16 unable to bind P-Top10, thus inhibiting
uptake of the solution by transpiration. One hour later all
transcription of
Fed-1.
Plant Biology: Petracek et al.
Proc. Natl. Acad. Sci. USA 95 (1998)
single VP16yTet line were subdivided in tissue culture during
long-term exposure (15). However, an external concentration
the plantlet stage to form clonal octuplets (see Methods)
of 1 mgyliter was ineffective in quickly repressing transcription
expressing Fed-1. Five independent lines then were used for
of the P-Top10::Fed-1 transgene. Although we eventually
each of the treatments indicated.
observed decay of the Fed-1 mRNA, the response was slow and
To ensure the seedlings grown on nylon membrane exhibited
inconsistent (ref. 20 and M.E.P., data not shown). Therefore,
a proper light response, we asked whether Fed-1 mRNA
to rapidly deliver sufficient amounts of tetracycline to the
abundance decreased after transfer to darkness. The abun-
plants, we increased the external concentration of tetracycline
dance of Fed-1 mRNA decreased approximately 3-fold after a
to 10 mgyliter. This concentration was used successfully for
24-hr dark treatment, indicating that these conditions were
half-life measurements in N. tabacum suspension cultures (17)
appropriate for measuring Fed-1 half-life (Fig. 2A). In addi-
and for rapid (0.5 hr) regulation of GUS by vacuum infiltration
tion, we asked what level of tetracycline was required to shut
in tobacco leaves (21).
off the P-Top10 promoter. Previously, 1 mgyliter of tetracy-
Because 10 mgyliter tetracycline potentially could affect
cytoplasmic translation and thus the light response of the Fed-1
cline has been shown to effectively repress P-Top10 during a
mRNA, we monitored the polyribosome profiles of total plant
mRNA in the presence and absence of tetracycline. In un-
treated plants we observed a significant shift from polyribo-
some fractions after only 1 hr of dark treatment, similar to what
has been seen in agar grown plants (Eric R. Hansen, personal
communication). Addition of tetracycline did not change the
total polyribosome profile from that seen in untreated plants
with the same light treatment (Fig. 2B), suggesting that the
concentration of tetracycline delivered to the plants does not
alter significantly overall translation. To further ensure that
tetracycline does not alter the normal Fed-1 mRNA disap-
pearance, we applied 10 mgyliter tetracycline to transgenic
plants containing the P35S::Fed-1 mRNA. As shown in Fig. 2A,
tetracycline had no effect on the normal dark decline of the
Fed-1 mRNA.
In addition to rapid delivery of tetracycline, we needed to
ensure rapid delivery of DCMU so we could observe the
effects of short-term inhibition of photosynthesis on Fed-1
mRNA half-life. We showed previously that Fed-1 mRNA is
dissociated from polyribosomes after a 40-hr incubation with
1 mM DCMU (8). Therefore, we monitored the Fed-1 mRNA
polyribosome profile 1 hr after delivery of 1 mgyml DCMU to
membrane-grown plants. Fig. 2C shows that within 1 hr after
DCMU treatment, the Fed-1 mRNA is dissociated from the
polyribosome fractions. Similar results were obtained for
Fed-1 mRNA in plants exposed to 1 hr of dark treatment (Eric
R. Hansen, personal communication). These data suggest that
inhibition of photosynthesis by DCMU rapidly alters the
polyribosome loading pattern of Fed-1 mRNA.
After establishing conditions that allowed rapid introduc-
tion of tetracycline and DCMU to intact, healthy plants, we
asked whether light regulation of Fed-1 mRNA abundance
occurs posttranscriptionally by a change in mRNA half-life. To
determine the half-life of Fed-1 mRNA we used
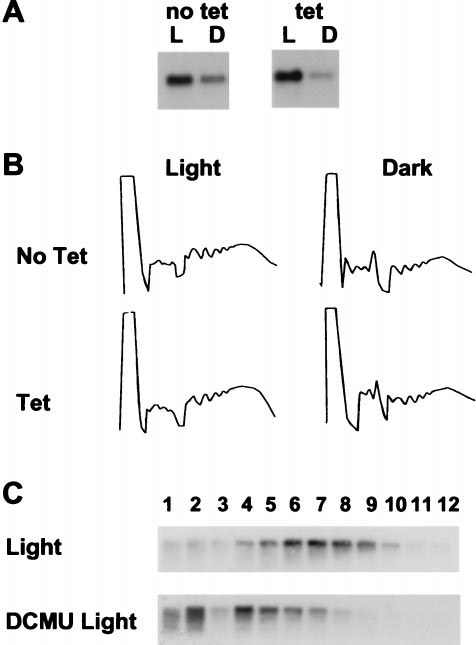
FIG. 2. Establishment of a hydroponic system to study Fed-1
P-Top10::Fed-1 transgenic tobacco seedlings (T2 generation)
mRNA half-life. (A) Decline of Fed-1 mRNA abundance in the dark.
grown on nylon membranes and isolated RNA samples at the
Fifteen to 20 transgenic nylon membrane grown seedlings from a
transgenic line containing P-Top10::Fed-1 treated with (tet) or without
times indicated in Fig. 3. To assay plants grown under different
tetracycline (no tet) were either left in the light for 24 hr (L) or
conditions, we simultaneously performed the same experi-
wrapped in foil for dark treatment for 24 hr from the time the light
ments on clonal plantlets. In both plant systems (see Methods),
sample was harvested (D). Total RNA extracted and 5 mg was
identical half-lives were observed.
separated in each lane of an agarose gel, blotted, and hybridized with
The half-life data from both types of growth systems are
32P-labeled Fed-1. (B) Lack of effect on total polyribosome loading by
combined and summarized in Fig. 3. For all three conditions,
tetracycline. Transgenic P35S::Fed-1 tobacco seedlings were treated
approximately the first 80% of the decay can be fit with a single
with either 10 mgyliter tetracycline in Hoagland's solution (Tet) or
first-order component, using the equation ln(CyC0) 5 2kdt
with Hoagland's solution alone (No Tet). After 1 hr of uptake the
and ln2ykd 5 t1/2, where C0 is the initial mRNA concentration,
plants were subjected to continued illumination (Light) or 1 hr of dark
treatment (Dark). Shown are absorbance tracings at 254-nm UV of the
C is the mRNA concentration at time t, kd is the decay
sucrose gradients containing extracts from these plants. The direction
constant, and t1/2 is the half-life of the mRNA (22). The
of the sedimentation is from left to right. (C) Effective DCMU uptake.
half-lives estimated for this major component are approxi-
Fifteen to 20 transgenic nylon membrane-grown seedlings from a
mately 2.4 hr for the light-treated plants and 1.2 hr for
transgenic line containing P-Top10::Fed-1 (nylon membrane grown)
dark-treated plants. Correlation coefficients are 0.989 and
either were not treated (Light) or treated with 1 mM DCMU (DCMU
0.999, respectively. The remaining 20% of the mRNA appears
Light) for 1 hr and then harvested into liquid nitrogen. RNAs from
to decay more slowly, although variation in the data prevents
each fraction of a sucrose gradient were resolved by gel electrophore-
accurate analysis of this minor component. The apparently
sis, blotted, and probed with antisense 32P-labeled RNA to either
slower decay might indicate a subset of the Fed-1 mRNA
Fed-1. Light to heavy sucrose fractions were loaded from left to right.
Amounts of hybridizing RNA cannot be quantitatively compared
decays more slowly than the bulk mRNA. However, measure-
between gradients because of variations in the amount of tissue used
ments in this range are based on weak hybridization signals and
and the efficiency of grinding.
may be influenced by background variations or a small amount
Plant Biology: Petracek et al.
Proc. Natl. Acad. Sci. USA 95 (1998)
and thus we could not answer whether the decline in mRNA
abundance was because of stress from a long-term dark
treatment or because of a more immediate, specific signal. The
immediate decrease in Fed-1 mRNA half-life after transfer of
the plants to darkness or treatment with DCMU suggests that
the response to the photosynthetic state of the plant is a result
of a rapid signal rather than a relatively slow change, such as
increases in abscisic acid (23).
Using the P-Top10 system, we can now more easily study
elements of the Fed-1 mRNA that affect its stability because
half-life measurements circumvent ‘‘position effect'' variation
in absolute expression levels among different transgenic lines.
It is difficult to firmly determine whether the stability of
mutant mRNAs is altered by comparing mRNA abundance
because transgene expression in plants can vary greatly. For
example, although we know the Fed-1 mRNA light response is
abolished when the AUG is mutated, we do not know whether
this results from increasing mRNA stability in the dark or
decreasing it in the light. Using the P-Top10 system, we will
now be able to make those determinations.
Previously, we showed that changes in Fed-1 mRNA abun-
dance are correlated with the ability of the mRNA to be
FIG. 3. The effect of photosynthesis on Fed-1 mRNA half-life.
translated (2). The data presented here support the suggestion
Fifteen to 20 control or DCMU-treated (1 mM) transgenic seedlings
that Fed-1 is preferentially degraded when it is not associated
per sample from a transgenic line containing P-Top10::Fed-1 (nylon
with polyribosomes. We propose that a signal generated by
membrane grown) or clonal octuplets were treated with 10 mgyliter
photosynthetic electron transport in the chloroplast travels to
tetracycline for 1 hr and then exposed to light (h), dark (r), or DCMU
the cytoplasm and signals a change in the cytoplasmic trans-
and light (E) for the time indicated on the x axis. Total RNA (5 mg)
lation rate of the Fed-1 mRNA. Furthermore, we suggest that
was separated in each lane of an agarose gel, blotted, and hybridized
a reduction in Fed-1 mRNA translation rate in the dark results
with 32P-labeled Fed-1. Blots were rehybridized with 32P-labeled
in a faster rate of degradation of the Fed-1 mRNA. Cis analysis
antisense HIS probe to detect the endogenous His HI transcript
of the Fed-1 59 UTR identified a region containing four
(divided square). The resulting hybridization signals were quantitated
by using a PhosphorImager (Molecular Dynamics). The mean percent
tandem copies of a CATT motif that is apparently sensitive to
reduction in RNA was calculated for each time point and plotted on
nuclease degradation (7). When this region is mutated, Fed-1
a semi-log plot. Each time point is derived from at least eight separate
mRNA accumulates to a similar level in both the light and the
experiments, with the error bars representing SEM. A line was drawn
dark. However, the mutated Fed-1 mRNA that is not degraded
at 50% mRNA remaining.
in the dark still accumulates in the nonpolyribosomal fractions.
We suggest that darkness inhibits translation at the level of
of residual transcription from the P-Top10. It is therefore
initiation and that wild-type but not mutant Fed-1 mRNA is
premature to draw any conclusions based on this minor
degraded in the absence of polyribosome association. The
component of the decay curve.
dark-induced decrease in Fed-1 mRNA half-life thus appears
We also asked whether short-term inhibition of photosyn-
to be the result of a two-step process. First, Fed-1 mRNA
thesis by DCMU treatment would show the same effect on
dissociates from polyribosomes and second, the Fed-1 mRNA
half-life as a brief dark treatment. As shown in Fig. 3, the decay
is degraded in the absence of polyribosome protection. It
curve for Fed-1 mRNA in the presence of DCMU is similar to
would be interesting to know whether there is in fact a ‘‘labile''
that for mRNA of dark-treated leaves. Fitting a first-order
factor required for Fed-1 degradation in the dark as inferred
component to the first 80% of the decay curve yields a half-life
from the stabilization of Fed-1 mRNA in the presence of ActD.
of 1.2 hr (correlation coefficient 5 0.978), a value indistin-
If so, what is the nature of this factor? For example, a labile
guishable from the half-life of Fed-1 mRNA in darkness.
nuclease may interact with the CATT repeat; in this case we
Finally, as a control, we asked whether tetracycline addition
would predict that degradation would be inhibited when
affected endogenous Histone HI mRNA levels. As expected,
expression of the nuclease is inhibited. Alternatively, it is
Histone HI RNA levels remained constant in the presence of
possible that a labile factor is required to inhibit translational
tetracycline, indicating that tetracycline does not inhibit global
initiation in darkness. In either case, it will be interesting to
transcription or turnover of other mRNAs. Taken together,
sort out the specificity of Fed-1 degradation as well as the
our data show that photosynthetic signals control the stability
photosynthesis-responsive signaling pathways involved in the
of Fed-1 mRNA.
regulation of Fed-1 mRNA half-life.
We acknowledge excellent technical assistance from Tyrone Hughes
and Tamyra Ravenel. We are also grateful to Dr. Mark Longtine for
Here we present direct evidence that Fed-1 mRNA is post-
critical review of the manuscript. Controlled environment plant
transcriptionally light-regulated at the level of mRNA stability.
growth space was provided by the Southeastern Plant Environment
Previously, we had only indirect evidence that did not distin-
Laboratory (Raleigh, NC). This project was supported by National
guish between altered stability and altered transcriptional
Institutes of Health Postdoctoral Fellowship Grant 1F32GM15510–01
elongation. Furthermore, in the absence of Fed-1 transcrip-
to M.P., and National Science Foundation Grant MCB-9507396 and
tion, a difference in Fed-1 mRNA abundance is detectable
National Institutes of Health Grant GM43108 to W.F.T. and L.F.D.
within an hour of the cessation of photosynthesis, suggesting
that the change in Fed-1 mRNA stability is rapid. Previously
1. Elliott, R. C., Dickey, L. F., White, M. J. & Thompson, W. F.
(1989) Plant Cell 1, 691–698.
we showed that the decrease in Fed-1 mRNA abundance is
2. Dickey, L. F., Nguyen, T. T., Allen, G. C. & Thompson, W. F.
correlated with a dissociation from polyribosomes in response
(1994) Plant Cell 6, 1171–1176.
to cessation of photosynthesis. However, we had not been able
3. Thompson, W. F., Everett, M., Polans, N. O., Jorgensen, R. A. &
to determine whether this response was immediate or delayed,
Palmer, J. D. (1983) Planta 158, 487–500.
Plant Biology: Petracek et al.
Proc. Natl. Acad. Sci. USA 95 (1998)
4. Kaufman, L. S., Roberts, L. R., Briggs, W. R. & Thompson, W. F.
14. Thompson, W. F. & White, M. J. (1991) Annu. Rev. Plant Physiol.
(1986) Plant Physiol. 81, 1033–1038.
Mol. Biol. 42, 423–466.
5. Gallo-Meagher, M., Sowinski, D. A., Elliott, R. C. & Thompson,
15. Gatz, C. (1995) Methods Cell Biol. 50, 411–424.
W. F. (1992) Plant Cell 4, 389–395.
16. Weinmann, P., Gossen, M., Bujard, H., Hillen, W. & Gatz, C.
6. Dickey, L. F., Gallo-Meagher, M. & Thompson, W. F. (1992)
(1994) Plant J. 5, 559–569.
EMBO J. 11, 2311–2317.
17. Gil, P. & Green, P. (1996) EMBO J. 15, 1678–1686.
7. Dickey, L. F., Petracek, M. E., Nguyen, T. T. & Thompson, W. F.
18. Elliott, R. C., Pedersen, T. J., Fristensky, B., White, M. J., Dickey,
(1998) Plant Cell 10, 475–484.
L. F. & Thompson, W. F. (1989) Plant Cell 1, 681–690.
8. Petracek, M. E., Dickey, L. F., Huber, S. C. & Thompson, W. F.
19. Horsch, R. B., Fry, F. E., Hoffman, N. L., Eicholtz, D., Rogers,
(1997) Plant Cell 9, 2291–2300.
9. Sullivan, M. L. & Green, P. J. (1996) RNA
S. G. & Fraley, R. T. (1985) Science 227, 1229–1231.
10. Sheu, J.-J., Jan, S.-P., Lee, H.-T. & Yu, S.-M. (1994) Plant J 5,
20. Petracek, M. E., Dickey, L. F., Hansen, E. R., Sowinski, D. A.,
Nguyen, T. T., Allen, G. C. & Thompson, W. F. (1998) in A Look
11. Fritz, C. C., Herget, T., Wolter, F. P., Schell, J. & Schreier, P. H.
Beyond: Mechanisms Determining mRNA Stability and Translation
(1991) Proc. Natl. Acad. Sci. USA 88, 4458–4462.
in Plants: Proceedings of 19th Annual Riverside Symposium in Plant
12. Zhang, S., Sheng, J., Liu, L. & Mehdy, M. C. (1993) Plant Cell 5,
Physiology, ed. Gallie, D. (Am. Soc. Plant Physiol., Rockville,
MD), pp. 96–101.
13. Petracek, M. E., Dickey, L. F., Nguyen, T., Allen, G. A., Sowinski,
21. Gatz, C., Kaiser, A. & Wendenburg, R. (1991) Mol. Gen. Genet.
D. A., Hansen, E. R. & Thompson, W. F. (1996) in Regulation of
Plant Growth and Development by Light, eds. Briggs, W. R.,
22. Abler, M. & Green, P. J. (1996) Plant Mol. Biol. 32, 63–78.
Heath, R. L. & Tobin, E. M. (Am. Soc. Plant Physiol., Rockville,
23. Weatherwax, S., Ong, M., Degenhardt, J., Bray, E. & Tobin, E.
MD), pp. 80–88.
(1996) Plant Physiol. 111, 363–370.
Source: http://www.ufv.br/dbv/pgfvg/BVE684/htms/pdfs_seminarios/seminario1/n%EDvel_2/express%E3o_genica/Ferredoxin1%20mRNA%20is%20destabilized%20by%20changes%20in%20photosyntheti.pdf
See discussions, stats, and author profiles for this publication at: ARTICLE in INTERNATIONAL JOURNAL OF LAW AND PSYCHIATRY · APRIL 2013Impact Factor: 1.19 · DOI: 10.1016/j.ijlp.2013.04.017 · Source: PubMed 4 AUTHORS, INCLUDING: 67 PUBLICATIONS 176 CITATIONS 2 PUBLICATIONS 0 CITATIONS Hope and Healing Center & Institute87 PUBLICATIONS 4,082 CITATIONS
Topic Introduction Immersion Freezing of Biological Specimens: Rationale, Principles,and Instrumentation Guenter P. Resch,1,5 Marlene Brandstetter,1 Angela M. Pickl-Herk,2 Lisa Königsmaier,3Veronika I. Wonesch,1,4 and Edit Urban3 1IMP-IMBA-GMI Electron Microscopy Facility, Institute of Molecular Biotechnology, 1030 Vienna, Austria2Max F. Perutz Laboratories, Medical University of Vienna, 1030 Vienna, Austria3Institute of Molecular Biotechnology, 1030 Vienna, Austria4University of Applied Sciences Wiener Neustadt, 2700 Wiener Neustadt, Austria

