Microsoft word - schuster_mrmc.doc
Mini-Reviews in Medicinal Chemistry, 2006, 6, 909-920
S-Layer Proteins as Key Components of a Versatile Molecular
Construction Kit for Biomedical Nanotechnology
B. Schuster*, D. Pum, M. Sára and U.B. Sleytr
Center for NanoBiotechnology, University of Natural Resources and Applied Life Sciences, 1180 Vienna, Austria
Abstract: Surface (S)-layer proteins and S-layer fusion proteins incorporating functional sequences, self-assemble into
monomolecular lattices on solid supports and on various lipid structures. Based on these S-layer proteins, supramolecular
assemblies can be constructed which are envisaged for label-free detection systems, as affinity matrix, as anti-allergic
immuno-therapeutics, as membrane protein-based screening devices, and as drug targeting and delivery systems.
Keywords: Crystalline surface layer proteins, artificial virus, biomimetics, bottom-up strategy, S-layer fusion protein,
microspheres-based detoxification system, nanobiotechnology.
2. GENERAL ASPECTS OF S-LAYER PROTEINS
The cross fertilization of biology, molecular biology,
Crystalline bacterial cell surface layers, referred to as S-
organic chemistry, material sciences, and physics has opened
layers [1-3] have now been identified in hundreds of different
up significant opportunities for innovation in previously
species of bacteria and represent an almost universal feature
unrelated fields. In this context, self-assembly is a new and
of archaea (Fig.
1) [for reviews see 2, 4-10]. Since S-layers
rapidly growing scientific and engineering discipline that
are composed of a single protein or glycoprotein species
crosses the boundaries of numerous existing fields. Self-
endowed with the ability to assemble into a monomolecular
assembly can be defined as a "bottom-up" process by which
lattice during all stages of cell growth and cell division, they
individual molecules (ranging in size up to large polymers)
can be considered as the simplest type of biological
become spontaneously organized into supramolecular
membranes developed in the course of evolution [for review
structures. This alternative to "top-down" processing steps
can lead to both, new materials and structures that are not
S-layers can be associated with quite different supporting
obtained by conventional techniques, and to the ultimate
supramolecular structures. In most archaea, S-layers
miniaturization of functional units.
represent the only wall component and can be so closely
One of the great challenges for nano(bio)technology is
associated with the plasma membrane that a hydrophobic
the creation of supramolecular materials in which the
domain of the constituent subunits is actually integrated into
constituent units are highly regular molecular nanostructures.
the lipid layer [6, 8, 12]. In most Gram-positive bacteria the
Thus, learning how to create complex and large supra-
S-layer is attached to a rigid wall matrix involving lectin
molecular structures and the elucidation of rules mediating
binding between a glycan (referred to as secondary cell wall
their organization into functional materials will offer a broad
polymer, SCWP) covalently-attached to the peptidoglycan
spectrum of new technologies.
meshwork [13]. In Gram-negative bacterial cell envelopes S-layers are linked to the lipopolysaccharide component of the
It is now well-recognized that crystalline bacterial cell
outer membrane. In most prokaryotic organisms S-layers
surface layers (S-layers) composed of identical protein-
have to be considered as non-conservative structures with the
aceous subunits represent unique patterning elements and
potential to fulfil a broad spectrum of functions [3, 4, 9].
scaffolding structures for nanobiotechnological applications.
Considering that S-layer carrying organisms are ubiquitous
The possibility for incorporating single or multifunctional
in the biosphere and even dwell under the most extreme
domains to S-layer proteins by genetic engineering has led to
environmental conditions, the supramolecular concept of a
ultimate control and precision in the spatial distribution and
dynamic closed crystalline surface layer could have the
orientation of molecules and functional domains as required
potential to fulfil a broad spectrum of functions. Because S-
for life- and non-life science applications. Most relevant, S-
layer lattices possess pores identical in size and morphology
layers represent the base for very versatile self-assembly
in the 2 to 8 nm range, they work as precise molecular sieves
systems involving all major species of biological molecules
providing sharp cut off levels for the bacterial cell [14]. As
such as proteins, lipids, glycans, nucleic acids, and
isoporous ultrafiltration membrane they can apparently
combination of that.
provide the microorganisms with a selection advantage byfunctioning as protective coats, molecule and ion traps, andas a structure involved in cell adhesion, surface recognition
*Address correspondence to this author at the Center for Nano-
or antifouling [5, 11, 12, 15]. In those archaea which possess
Biotechnology, University of Natural Resources and Applied Life Sciences
S-layers as exclusive envelope component outside the
Vienna, Gregor-Mendel-Strasse 33, 1180 Vienna, Austria; Tel: ++43-1-47654-2200; Fax: ++43-1-4789112; E-mail:
[email protected]
cytoplasmic membrane, the crystalline array acts as a frame
2006 Bentham Science Publishers Ltd.
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
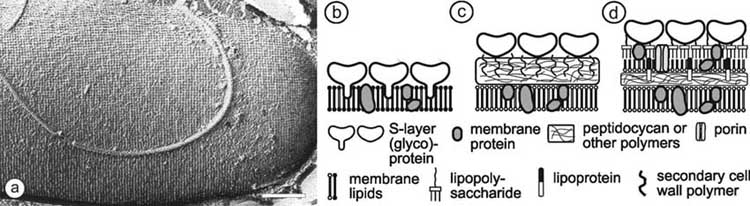
Fig. (1). In (a), freeze-etching preparation of a whole cell of Bacillus sphaericus with a square S-layer lattice is shown. Bar corresponds to
200 nm. Schematic illustration of the supramolecular architecture of the three major classes of prokaryotic cell envelopes containing
crystalline bacterial cell surface layers (S-layers). (b) Cell envelope structure of gram-negative archaea with S-layers as the only component
external to the cytoplasmic membrane. (c) Cell envelope as observed in gram-positive archaea and bacteria. In bacteria the rigid wall
component is primarily composed of peptidoglycan. In archaea other wall polymers (e.g. pseudomurein) are found. (d) Cell envelope profile
of gram-negative bacteria composed of a thin peptidoglycan layer and an outer membrane. If present, the S-layer is closely associated with
the lipopolysaccharide of the outer membrane. Modified after Ref. [5], Copyright (1999) with permission from Wiley-VCH.
work that determines and maintains the cell shape and
Contrary to the electron microscopical preparation tech-
stabilizes the cytoplasmic membrane [16, 17].
niques, scanning force microscopy allows to investigate S-layer monolayers in their native environment [26-28].
From a general point of view S-layers as the most
Contact mode microscopy in liquid is most frequently used
abundant of bacterial cellular proteins are important model
to investigate S-layer protein monolayers at sub-nanometer
systems for studying the structure, synthesis, assembly, and
resolution. S-layer proteins are highly susceptible towards
function of these proteinaceous components. The investi-
the applied tip loading forces which shall not exceed 0.5 to 1
gation of the general principles of S-layers also have
nN. Ionic content and strength of the buffer solution in the
revealed considerable application potential in biotechnology,
liquid cell has to be carefully adjusted in order to minimize
biomimetics, and nano(bio)technology [11, 15, 18-21].
electrostatic interactions between tip and sample. Silicon
2.1. Structural Analysis of S-Layer Lattices
wafers and mica are the most commonly used substrates forscanning force microscopical investigations since these
High resolution transmission electron microscopy (TEM)
provide hard and very flat surfaces. In particular, silicon
and scanning force microscopy (SFM) are commonly used to
surfaces are most relevant for nanobiotechnological
characterize S-layer protein lattices. In particular, in TEM
applications. S-layer proteins recrystallize into large scale
the appropriate preparation method is most important for
monomolecular protein lattices on silicon, whereas S-layer
investigating the ultrastructure of S-layer protein lattices at
fragments or self-assembly products are preferably deposited
molecular resolution (Fig. 1). Freeze-etching and freeze-
on mica. If S-layer proteins are recrystallized on flat solid
drying in combination with heavy metal shadowing are the
supports such as silicon wafers, lattice formation can be
most straight forward approaches for obtaining information
followed in real time [26]. It could be demonstrated that
about the lattice type and surface structure of S-layers on
crystal growth starts at several distant nucleation points and
bacterial cells and S-layer cell wall fragments [1, 22]. These
proceeds in-plane until a closed layer of crystalline domains
studies revealed that many S-layers show a smooth outer and
is formed [26]. The scanning force microscope has been also
a more corrugated inner face [23, 24]. This difference is of
used as a nano-tool for inducing conformational changes in
particular importance when the orientation (sidedness due to
S-layer proteins [29, 30]. Furthermore, the capability of
attachment of the S-layer subunits via the inner or outer
scanning force microscopy to resolve molecular details on
surface) of S-layers on artificial substrates has to be deter-
biological samples together with its force detection
mined. Nevertheless, TEM of frozen hydrated specimens
sensitivity has led to the development of the so-called
[23-25] yields the highest resolution among all microscopical
"topography and recognition mode", a method suitable for
techniques. In plane, a resolution of 0.35 nm and in the third
visualizing the chemical composition of a sample while
dimension 0.7 nm is attainable. In three-dimensional TEM,
mapping its topography [31]. It is anticipated that the
tilt series of the specimen is recorded under low electron
simultaneous investigation of both, topography and
dose conditions (usually not more than 1 to 2 electrons per
recognition, will allow to elucidate the structure-function
Å2). Although quantum noise governs image formation, such
relationship of a broad spectrum of biological samples in an
low electron doses are mandatory in order to maintain the
unsurpassed way.
three-dimensional structure of the proteins [25]. Imageprocessing methods are used to enhance the signal-to-noise
2.2. Self-assembly Properties of S-Layer Proteins
ratio in low dose micrographs.
While many archaeal S-layer proteins are covalently
Negative staining is an easy preparation technique in
anchored, those of bacteria are non- covalently linked to
TEM. Particularly in combination with two and three dimen-
each other and to the supporting cell wall component. Thus,
sional image reconstruction techniques, it allows high reso-
a complete solubilization of S-layers into their constituent
lution studies of the ultrastructure of S-layer lattices [23-25].
subunits and release from the bacterial cell envelope can be
S-Layer Proteins as Key Components
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8 911
achieved by treatment with high concentrations of hydrogen-
25 mol % are charged amino acids, and S-layer proteins
bond breaking agents (e. g. urea, guanidinium hydro-
possess little or no sulfur-containing amino acids. Secondary
chloride), by dramatic changes in the pH-value or in the salt
structure predictions of S-layer proteins indicate that about
concentration. Upon removal of the disrupting agent, e. g. by
40 % occur as �-sheets and approximately 20 % of the amino
dialysis, S-layer proteins self-assemble into two dimensional
acids are organized as �-helices. Most �-helical segments
arrays [for review see ref. 32]. Such self-assembly products
are arranged in the N-terminal part. Aperiodic foldings and
may have the form of flat sheets or open-ended cylinders.
�-turn content may vary between 5 % and 45 %.
Depending on the particular S-layer protein species used and
In order to elucidate the structure-function relationship of
on the environmental conditions, monolayers or double
distinct segments of S-layer proteins, N- and / or C-
layers are formed.
terminally truncated forms were produced and their self-
Contrary to the reassembly in solution, prior to
assembly and recrystallization properties investigated [44-
recrystallization on artificial supports, S-layer proteins must
46]. Another approach was seen in performing a cysteine
be kept in a water soluble state. This can either be achieved
scanning mutagenesis and screening the accessibility of the
in the absence of bivalent cations [33] or by maintaining a
single introduced cysteine residue in the soluble, self-
sub-critical protein concentration for self-assembly [34]. In
assembled and recrystallized S-layer proteins [34]. This
addition, in the presence of S-layer-specific SCWPs, the
study elucidated which amino acid positions in the primary
reassembly in suspension is inhibited, whereas the recrystalli-
sequence are located on the outer or inner S-layer surface of
zation of soluble S-layer proteins on artificial supports is
the subunits, inside the pores, or at the subunit to subunit
promoted [13, 34, 35]. The formation of coherent crystalline
arrays strongly depends on the S-layer protein species, the
The fact that no structural model at atomic resolution of
environmental conditions of the bulk phase (e. g. temperature,
an S-layer protein is available until now, may be explained
pH-value, ion composition and ionic strength) and, in parti-
by the molecular mass of the subunits being too large for
cular, on the surface properties of the substrate. For example,
nuclear magnetic resonance analysis, as well as by the
the S-layer protein SbpA of Bacillus sphaericus CCM2177
intrinsic property of S-layer proteins to self-assemble into
forms double layers with perfect long range order (up to
two dimensional lattices, thereby hindering the formation of
several micrometers in diameter) on hydrophilic silicon but
isotropic three dimensional crystals as required for X-ray
monolayers consisting of 200 to 500 nm sized patches on
crystallography. In addition, the low solubility of S-layer
proteins is a general hindrance for both methods.
In accordance with S-layer proteins recrystallized on
In the case of the S-layer protein SbsC of G .
solid substrates, the orientation of the protein arrays
stearothermophilus ATCC 12980, water soluble N- or C-
(sidedness due to the attachment via the inner or outer
terminally truncated forms were used for first three dimen-
surface of the S-layer subunits) at liquid interfaces and at
sional crystallization studies. Crystals of the C-terminally
lipid films is determined by the anisotropy in the physico-
chemical surface properties of the protein lattice [for review
31-844 diffracted to a resolution of 3 Å using
synchrotron radiation [47]. Native and heavy atom derivative
see ref. 36]. For example, the S-layer protein SbsB of
data confirmed the results that the N-terminal region is
Geobacillus stearothermophilus pv72/p2 reassembles with
mainly organized as �-helices, whereas the middle and C-
its more hydrophobic outer face at the air-water interfaces
terminal part of SbsC consist of loops and ß-sheets [47].
while at lipid films with zwitterionic head groups the S-layerlattice is attached with its inner face [37]. The unambiguous
The N-terminal region was found to be responsible for
determination of the orientation of the S-layer is possible
anchoring the S-layer subunits to the underlying rigid cell
since it shows oblique lattice symmetry with a characteristic
envelope layer by binding to the SCWP. The polymer chains
handedness of the proteins. In addition to the formation of
are covalently linked to the peptidoglycan backbone which
flat S-layer lattices it has also been demonstrated that S-layer
occurs most probably via phosphodiester bonds [48].
proteins are able to cover liposomes and nanocapsules
Basically, two types of binding mechanisms between the N-
completely [38-49]. The S-layer shows facets and numerous
terminal part of S-layer proteins and SCWPs have been
lattice faults in order to follow the curvature of the spheres.
described [49]. The first one, which involves so-called S-
According to the observations with planar lipid films, the
layer-homologous (SLH) domains and pyruvylated SCWPs
charge of the lipid head groups and the polyelectrolyte
[33, 40, 44, 50, 51] has been found to be widespread among
determines the orientation of the S-layer protein against the
prokaryotes and is considered as having been conserved in
liposome and the nanocapsule, respectively [41, 42].
the course of evolution [51]. The second type of bindingmechanism has been described for G. stearothermophilus
2.3. Chemical Properties and Molecular Biology of S-
PV72/p6 and ATCC 12980 [9, 52, 53], a temperature-derived
strain variant from the latter [54], and G. stearothermophilus
Chemical analyses and genetic studies revealed that the
NRS 2004/3a [55]. This binding mechanism involves an
S-layer lattices are composed of a single homogeneous
SCWP that consists of N-acetyl glucosamine, glucose and
protein or glycoprotein species with a molecular mass
2,3-dideoxy-diacetamido-D-mannosamine uronic acid in the
ranging from 40 to 200 kDa [5, 9, 11, 43]. Most S-layer
molar ratio of 1:1:2 (see compound (1) in Fig. 2) [55, 56]
proteins are weakly acidic with isoelectric points in the range
and a highly conserved N-terminal region which does not
of 4 to 6 [9]. In general, S-layer proteins consist of a large
possess an SLH-domain [52-55]. Concerning the first
portion of hydrophobic amino acids (40 - 60 mol %), about
binding mechanism, the construction of knock-out mutants
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
Fig. (2). Chemical structure of the repeating unit of the secondary cell wall polymer of G. stearothermophilus NRS 2004/3a (1).
in Bacillus anthracis and Thermus thermophilus in which the
parameters of a = 10.4 nm, b = 7.9 nm and a base angle of �
gene encoding a putative pyruvyl transferase was deleted
= 81°, whereas SbpA assembles into a square lattice with a
demonstrated that the addition of pyruvic acid residues to the
lattice constant of 13.1 nm.
peptidoglycan-associated cell wall polymer was a necessary
For generating a universal affinity matrix for binding any
modification to bind SLH-domain containing proteins [50,
kind of biotinylated molecule, S-layer-streptavidin fusion
proteins were constructed [60, 61]. Minimum-sized core
3. A MOLECULAR CONSTRUCTION KIT BASED ON
streptavidin (118 amino acids) was either fused to N- or C-
terminal positions of SbsB or to the C-terminal end ofrSbpA31-1068 [45, 61].
The biomimetic approach learning from nature how to
create supramolecular, layered structures by a bottom-up
The genes encoding the fusion proteins and core
process is one of the most challenging scientific tasks in
streptavidin were expressed independently in E. coli and
nanobiotechnology. Advantage can be taken of the self-
isolated from the host cells. To obtain functional hetero-
assembling nature of S-layer (fusion) proteins, SCWPs and
tetramers (HTs), a refolding procedure was developed by
natural and/or artificial lipids and their properties in the
subjecting a mixture of fusion protein with excess core
compartmentalization of components in nanoscale regions,
streptavidin to denaturing and renaturing conditions and
production of self-assembling biomaterials, construction of
isolating functional HTs by size exclusion and affinity
drug-targeting and delivery systems, and development of
chromatography. HTs comprising the N-terminal rSbsB-
smart biosensors [18, 19, 57-59].
streptavidin formed self-assembly products in suspensionand recrystallized on liposomes and silicon wafers [61],whereas HTs based on the C-terminal rSbpA
3.1. S-Layer Fusion Proteins and their Application
vidin fusion protein showed dirigible self-assembly formation,
as lattice formation of SbpA is strongly dependent on the
So far, the chimaeric genes encoding several S-layer
presence of calcium ions. HTs based on the rSbpA31-1068-
fusion proteins have been heterologously expressed in
streptavidin fusion protein recrystallized on gold surfaces
Escherichia coli. S-layer fusion proteins were based on the
that were optionally pre-coated with SCWP [60]. Analysis of
S-layer proteins SbsB, SbsC, and SbpA (Table 1). SbsB
negatively-stained preparations of self-assembly products
forms an oblique S-layer lattice with p1 symmetry and lattice
formed by HTs revealed that neither the oblique S-layer
Summary of Various S-Layer Fusion Proteins (Selected from Various Constructs)
S-layer fusion protein
Length of functionality
rSbsB / core streptavidin
/ core streptavidin
major birch pollen allergen
rSbpA / Strep-tag I
affinity tag for streptavidin
IgG-binding domain
green fluorescent protein
heavy chain camel antibody
Mature proteins: Bacillus sphaericus CCM2177 variant A (SbpA): 1238 amino acids (aa); Geobacillus stearothermophilus pv72/p2 (SbsB): 889 aa; Bacillus stearothermophilusATCC 12980 (SbsC): 1099 aa.
S-Layer Proteins as Key Components
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8 913
lattice of SbsB, nor the square lattice of SbpA had changed
monoclonal antibodies that recognize free, as well as PSA
due to the presence of the fusion partner. Digital image
complexed with alpha-1-anti-chymotrypsin. For application
reconstructions of self-assembly products of HTs comprising
in a PSA biosensor, VHHs recognizing free and complexed
the N-terminal rSbsB-streptavidin fusion protein showed an
PSA are desired. Moreover, kinetic requirements in the
additional protein mass on the N-terminal SLH-domain
biosensor impose a high probe density that can probably only
which resulted from the fused streptavidin moiety [61]. As a
be obtained with single domain VHHs.
first application approach, monolayers of HTs based on
To generate a PSA-specific sensing layer for SPR
rSbpA31-1068 were recrystallized on plain gold chips and on
measurements, the S-layer fusion protein rSbpA
those pre-coated with thiolated SbpA-specific SCWP, and
PSA-N7 was recrystallized on gold chips pre-coated with
the obtained affinity matrix was used to perform hybri-
thiolated SCWP. The formation of the monomolecular
dization experiments. In a first step, biotinylated oligo-
protein lattice was confirmed by scanning force microscopy,
nucleotides (30-mers) were bound to the streptavidin moiety
as well as by the level of the measured SPR signal. As
of the HTs, and complementary oligonucleotides were
derived from response levels measured for binding of PSA to
hybridized carrying no or one mismatch [60]. Evaluation of
a monolayer consisting of rSbpA
the hybridization experiments was performed by applying
31-1068 /cAb-PSA-N7, the
molar ratio between bound PSA and the S-layer fusion
protein was 0.78, which means that at least three PSA
which combines the advantages of the high optical field
molecules were bound per morphological unit of the square
intensities of surface plasmon waves with the sensitive
S-layer lattice with an area of 170 nm2. To summarize, by
detection of fluorescence light emission. For hybridization
using SbpA-specific SCWP as biomimetic linker to gold
experiments on monolayers generated by recrystallization of
chips, a sensing layer for SPR could be generated by
HTs on gold chips pre-coated with thiolated SCWP, fluo-
recrystallization of this S-layer fusion protein. Due to the
rescently labelled oligonucleotides carrying one mismatch
crystalline structure of the S-layer lattice, the fused ligands
were used. The fluorescence intensity increased linearly at
showed a well defined distance in the protein lattice, and
the beginning of the hybridization reaction, so that the linear
according to the selected fusion site in the S-layer protein,
slope of the increase in the fluorescence intensity plotted
they were located on the outermost surface, which should
versus the concentration of the hybridizing oligonucleotides
reduce diffusion limited reactions. A further advantage can
led to a linear correlation [60]. In a different set of
be seen in the constant and low distance of the ligands from
hybridization experiments which were performed on
the optically active gold layer, which is exclusively
monolayers generated by direct recrystallization of HTs on
determined by the thickness of the S-layer and lies in the
plain gold chips, the concentration of oligonucleotides
range of only 10 to 15 nm. Thus, S-layer fusion proteins
carrying one mismatch was step-wise increased. The
incorporating camel antibody sequences can be considered as
Langmuir isotherm which indicated that oligonucleotides in
key element for the development of label free detection
solution were in equilibrium with those bound to the
systems such as SPR, surface acoustic wave, or quartz
monolayer carrying the biotinylated oligonucleotides could
crystal microbalance, in which the binding event can be
be established from the obtained fluorescence intensities
measured directly by the mass increase without the need of
[60]. The detection limit was found to be 1.57 pM on
any labelled molecule.
monolayers generated by recrystallization of HTs on goldchips pre-coated with thiolated SCWP, whereas on plain
The sequence encoding rSbpA31-1068 was also used as
gold chips, the detection limit was determined to be at least
base form for the construction of an IgG-binding fusion
8.2 pM. To conclude, the hybridization experiments
protein [64]. As fusion partner, the sequence encoding the Z-
indicated that a functional sensor surface could be generated
domain, a synthetic analogue of the IgG-binding domain of
by recrystallization of HTs on gold chips, which could find
Protein A from Staphylococcus aureus, was used. To
numerous applications in (nano)biotechnology.
generate the S-layer fusion protein, the 5´-end of thesequence encoding two copies of the Z-domain was fused via
An S-layer fusion protein comprising the C-terminally
a short linker to the gene encoding rSbpA
truncated form rSbpA
31-1068 and the variable region of a
heterologous expression in E. coli, the S-layer fusion protein
heavy chain camel antibody directed against lysozyme was
was isolated from the host cells, purified by size exclusion
constructed. The Camelidae is the only taxonomic family
chromatography under denaturing conditions, dialysed and
known to possess functional heavy chain antibodies lacking
recrystallized on gold chips which were pre-coated with
light chains and the first constant region. These unique
thiolated SbpA-specific SCWP. As shown by scanning force
antibody isotypes interact with the antigen by virtue of a
microscopy, a monomolecular protein lattice with square
single variable domain, termed VHH. A single VHH domain
symmetry was formed. Native monolayers or monolayers
has a molecular mass of only 15,000 and is the smallest
cross-linked with the bifunctional imidoester dimethyl-
known complete antigen binding fragment from a functional
pimelimidate (DMP, compound (2)) (Fig. 3) were finally
immunoglobulin. As proof-of-principle was provided with a
exploited for binding of human IgG. The amount that could
fusion protein comprising a VHH directed against lysozyme
be bound by the native monolayer was 2.9 x 10-5 nM or 4.35
[62], an S-layer fusion protein incorporating the sequence of
ng IgG / mm2, whereas in the case of the DMP-cross-linked
a variable domain of a heavy chain camel antibody (cAb-
monolayer (Fig. 3) 2.8 x 10-5 nM or 4.20 ng IgG / mm2 could
PSA-N7) directed against the prostate-specific antigen (PSA)
attach. These values corresponded to 65 and 67 % of the
was constructed [63]. PSA is a useful marker to screen
theoretical saturation capacity of a planar surface for IgG
potential prostate cancer patients. The current diagnostic test
(6.5 ng / mm2) with the Fab regions occurring in the
systems determine the concentration of total PSA with
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
The major birch pollen allergen Bet v1 shares IgE
epitopes with all tree pollen allergens from closely related
species (e. g. alder hazel, hornbeam, beech). Because of highsequence identities among these allergens and well studied
cross-reactions with B-cell epitopes, Bet v1 represents a
model allergen. The gene encoding the chimaeric S-layerproteins rSbsC31-920/Bet v1 [64] and rSbpA31-1068/Bet v1 [45]
carrying Bet v1 at the C-terminal end were cloned and
expressed in E. coli. In a recent study, the applicability of
rSbsC31-920/Bet v1 as a novel approach to design vaccines
cross-linked S-layer protein rSbpA (3)
with reduced allergenicity in combination with strongimmune-modulating capacity for immunotherapy of type I
allergy could be demonstrated [67]. This fusion proteinexhibited all relevant Bet v1-specific B and T cell epitopes,
Fig. (3). Cross-linking of the recombinant S-layer protein rSbpA
but was significant less efficient in releasing histamine than
with the homobifunctional imidoester dimethylpimelimidate (DMP;
free Bet v1. In cells of birch pollen-allergic individuals, the
compound (2)). The spacer arm length of DMP is 0.92 nm.
fusion protein was capable of modulating the allergen-specific Th2-dominated response into a more balanced Th1/
condensed state. As derived from these binding capacities,
Th0-like phenotype accompanied by enhanced production of
on average 2.7 and 2.6 IgG molecules were bound per
IFN-� and IL-10. To conclude, rSbsC31-920/Bet v1 could
morphological unit of the square S-layer lattice consisting of
find application as carrier/adjuvants to design vaccines for
four identical subunits of the S-layer fusion protein. For
specific immunotherapy of type 1 allergy with improved
preparing biocompatible microparticles for the microspheres-
efficacy and safety [67].
based detoxification system (MDS) [65] to remove auto-antibodies from patients´ sera suffering from auto-immune
The nucleotide sequence encoding enhanced green
disease, the S-layer fusion protein was recrystallized on
fluorescent protein (EGFP), a red-shifted green fluorescent
SCWP-coated, 3 �m large cellulose-based microbeads (Fig.
protein (GFP)-derivative possessing a 30 times brighter
4). The MDS is an alternative approach to conventional
fluorescence intensity at 488 nm than wild-type GFP was
immunoadsorption systems, in which the plasma does not
fused to the 3´end of the sequence encoding the C-terminally
perfuse on an adsorption column, but is recirculated into a
truncated form rSbpA31-1068 [68]. The chimaeric gene
filtrate compartment of a membrane module. The addition of
encoding rSbpA31-1068/EGFP was expressed in E. coli,
microbeads to the plasma circuit would allow the rapid
whereby expression at 28°C instead of 37°C resulted in
removal of the pathogenic substrates. In the case of
clearly increased fluorescence intensity, indicating that the
microbeads that were covered with a native monolayer, the
folding process of the EGFP moiety was temperature
binding capacity was 1,065 �g human IgG / mg S-layer
sensitive. Comparison of excitation and emission spectra of
fusion protein. For DMP-treated microbeads, a binding
rEGFP and rSbpA31-1068/EGFP indicated identical maxima at
capacity of 870 �g IgG / mg S-layer fusion protein was
488 and 507 nm, respectively. Furthermore, this fusion
determined. These values corresponded to 78 or 65 % of the
protein was used for recrystallization on silicon wafers
theoretical saturation capacity of a planar surface for IgG
covered with polyelectrolytes, as well as for coating of
having the Fab regions in the condensed state. Bound IgG
hollow polyelectrolyte capsules. Fluorescence spectroscopy
could be eluted with glycine-HCl buffer at a pH value of 2.2
confirmed that the adsorption of rSbpA31-1068/EGFP on
and the microbeads were used for further IgG-binding
hollow capsules did not shift the fluorescence emission of
experiments [64].
the chromophore [41]. Finally, the recrystallization of this
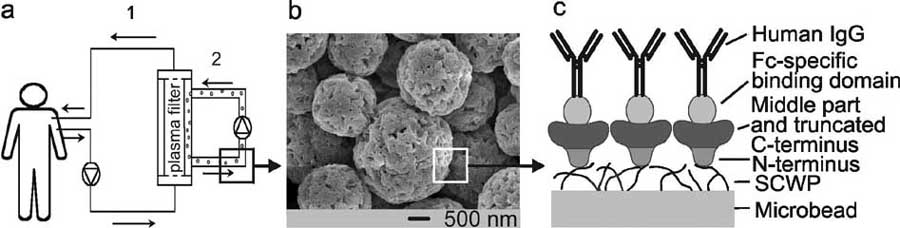
Fig. (4). (a) Schematic drawing of the MDS, showing the primary circuit (labelled 1) containing the whole blood of the patient. The blood
cells are rejected by the plasma filter. In the second circuit (labelled 2), the plasma re-circulates together with the S-layer fusion protein-
coated microbeads, on which IgG is bound. After passing the plasma filter again, the purified plasma is combined with blood cells, and the
whole blood is re-infused into the patient. (b) Scanning electron micrograph of the cellulose-based microbeads used for recrystallization of
rSbpA31-1068/ZZ. (c) Schematic drawing showing the oriented recrystallization of the S-layer fusion protein rSbpA31-1068/ZZ on microbeads
pre-coated with SCWP and binding of IgG to the ZZ-domains. Reprinted with permission from Ref. [64], Copyright (2004) ASM.
S-Layer Proteins as Key Components
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8 915
fusion protein on liposomes and their application is
exploited for covalent binding of functional macromolecules,
described in the following section.
like biotinylated antibodies v i a the streptavidin – biotinbridge [40]. These immuno-S-liposomes comprise several
3.2. S-Layer Fusion Proteins on Liposomes as Model for
components with specific functions: the liposome as drug
carrier, the antibody as homing device, the S-layer lattice as
Biomolecular self-assembly can be used as powerful tool
stabilizing structure for the liposome, as anchoring layer for
for nanoscale engineering. One well known example is the
the antibodies, and most probably as stealth coat for
formation of liposomes, which are still very promising supra-
prolonged blood circulation times (Fig. 5).
molecular structures for the application in nanobiotechno-
To avoid chemical modification reactions and to prevent
logy and nanobiomedicine.
diffusion of potentially toxic agents through the lipid bilayer
Liposomes are colloidal, vesicular structures based on
into the interior of the vesicles, S-layer fusion proteins
(phospho)lipid bilayers or on tetraetherlipid monolayers [69]
incorporating the sequence of core–streptavidin have been
and they are widely used as delivery systems for enhancing
constructed. Functional streptavidin HTs were prepared as
the efficiency of various biologically active molecules and
three of the four binding pockets remained accessible for
for the transport of therapeutic agents to the site of disease in
binding biotinylated molecules [61]. After recrystallization
vivo [70, 71]. Liposomes can encapsulate water soluble
of this streptavidin fusion protein on positively charged
agents in their aqueous compartment and lipid soluble
liposomes, the protein lattice was further functionalized by
substances within the lipid bilayer itself [72]. These agents
binding biotinylated peroxidase or biotinylated ferritin [61].
include small molecular drugs used in cancer chemotherapy
Binding of biotinylated ligands to S-liposomes can be
and genetic drugs as plasmids encoding therapeutic genes
exploited for enabling receptor-mediated uptake into human
[73]. Generally, liposomes release their contents by
cells. A further promising application potential can be seen
interaction with target cells, either by adsorption,
in the development of drug targeting and delivery systems
endocytosis, lipid exchange or fusion [71, 74].
based on lipid-plasmid complexes coated with functional
In previous studies, wild-type SbsB has been recrystalli-
HTs for transfection of human cells.
zed on positively charged liposomes composed of dipalmi-
Another interesting approach can be seen in the
toylphosphatidylcholine, cholesterol and hexadecylamine
generation of a functional chimaeric rSbpA31-1068/EGFP
[38-40, 75]. Such S-layer-coated liposomes (S-liposomes)
fusion protein to follow the uptake of S-liposomes into
with a diameter of 50–200 nm represent simple model
mammalian cells [68]. Liposomes coated with a monolayer
systems resembling the architecture of artificial virus
of rSbpA31-1068/EGFP were incubated with HeLa cells.
envelopes (Fig. 5). For that reason, S-liposomes could reveal
Subsequently, confocal laser scanning microscopy was
a broad application potential, particularly as drug delivery
applied to investigate the ongoing interaction between the
systems or in gene therapy [5].
fluorescently labelled cell membrane and the greenfluorescent S-liposomes. This study demonstrated that mostof the S-liposomes were internalized within 2 hours ofincubation and that the major part entered the HeLa cells byendocytosis [68]. To our knowledge, rSbpA31-1068/EGFP isthe first fusion protein that maintained the ability tofluorescence and to recrystallize into a monomolecularprotein lattice. Due to the intrinsic fluorescence, liposomescoated with rSbpA31-1068/EGFP represent a useful tool tovisualize the uptake of S-liposomes into mammalian cells.
The most interesting advantage can be seen in therecrystallization of fusion proteins incorporating EGFP incombination with HTs on the same liposome surface. In that
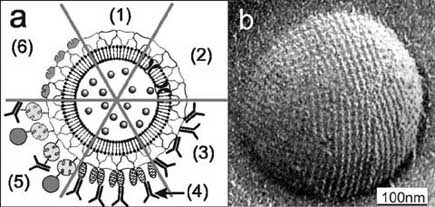
Fig. (5). (a) Schematic drawing of (1) an S-liposome with
case, it would be possible to simultaneously investigate the
entrapped functional molecules and (2) functionalized by
uptake of these specially coated S-liposomes by target cells
reconstituted integral proteins. S-liposomes can be used as
and the functionality of transported drugs without the
immobilization matrix for functional molecules (e.g. IgG) either by
necessity of additional labelling procedures.
direct binding (3), by immobilization via the Fc-specific ligandprotein A (4), or biotinylated ligands can be bound to the S-
3.3. S-Layer Based Lipid Chips
liposome via the biotin–streptavidin system (5). (6) Alternatively,
Biological membranes have attracted lively interest, as
liposomes can be coated with genetically modified S layer proteins
the advances in genome mapping revealed that
incorporating functional domains. (b) Electron micrograph of a
approximately one-third of all genes in an organism encode
freeze-etched preparation of an S-liposome. Bar: 100 nm. Reprinted
membrane proteins, such as ion channels, receptors, and
with permission from Ref. [15], Copyright (2002) Wiley-VCH.
membrane-bound enzymes [76]. In addition, more than 60 %
S-liposomes possess significantly enhanced stability
of all consumed drugs act on membrane proteins [77].
towards thermal and mechanical stress factors [39]. For
Therefore, the generation of stabilized lipid membranes with
generating targeted S-liposomes, the S-layer lattice on
functional membrane proteins represents a challenge to apply
liposomes was cross-linked with bis(sulfosuccinimidyl)
membrane proteins as key elements in drug discovery,
protein-ligand screening and biosensors.
3; compound (4 )) (Fig. 6), biotinylated and

Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
cross-linked S-layer protein SbsB (5)
Fig. (6). Cross-linking of the S-layer protein SbsB with the water-soluble, homobifunctional N-hydroxysuccinimide-ester bis(sulfosucci-
nimidyl) suberate (BS3; compound (4)). The spacer arm length of BS3 is 1.14 nm.
S-layer-supported lipid membranes (SLM) mimic the
supramolecular assembly of archaeal cell envelope
structures, as they are composed of a cytoplasmic membrane
and a closely associated S-layer [36]. In this biomimetic
architecture, either a tetraetherlipid monolayer, or an
artificial phospholipid bilayer replaces the cytoplasmic
membrane and isolated bacterial S-layer proteins are
attached either on one or both sides of the lipid membrane
(Fig. 7). The most commonly used lipids to generate planar
SLMs are the phospholipid 1,2-diphytanoyl-sn-glycero-3-
phosphatidylcholine (6), and the membrane-spanning tetra-
etherlipids Main Phospholipid (7) isolated from Thermo-
plasma acidophilum and glycerol dialkyl nonitol tetra-
Fig. (7). Schematic drawing of an S-layer covered (modified) solid
etherlipid (8) extracted and purified from Sulfolobus and
support (e.g. a gold electrode) carrying a lipid bilayer generated by
Metallosphaera archaea (Fig. 8).
vesicle fusion or by the Langmuir-Blodgett-technique. Integral
Electrostatic interactions between exposed carboxylic
membrane proteins can be reconstituted into this SLM. Further-
acid groups on the inner surface of the S-layer lattice and the
more, a second S-layer lattice can be recrystallized on the top of
zwitterionic lipid head groups were found to be primarily
this biomimetic structure to provide an enhanced long-term stability
responsible for the defined binding of the S-layer subunits.
and to act as a protective coat with pores in the nanometer range.
As two to three contact points between the lipid film and theattached S-layer protein were determined, only few lipid
In reconstitution experiments, the self-assembly of the
molecules were anchored to protein domains on the S-layer
staphylococcal pore-forming protein �-hemolysin (�HL)
lattice having a unit cell with a spacing of about 8 to 13 nm
[86] was examined at plain and SLMs [87]. �HL forms lytic
[78]. The remaining scores of lipid molecules diffused freely
pores when added to the lipid-exposed side of the S-layer-
in the membrane between the pillars consisting of anchored
supported membrane. No assembly was detected upon
lipid molecules. Because of its widely retained fluid
adding �HL monomers to the S-layer-face of the composite
characteristics, this nano-patterned lipid membrane was
membrane. Therefore, it was concluded that the intrinsic
termed "semifluid membrane" [79]. But most important,
molecular sieving properties of the S-layer lattice did not
although peptide side groups of the S-layer protein
allow the formation of �HL heptamers within the S-layer
interpenetrated the phospholipid head group regions almost
pores. Most interestingly, in SLMs the attached S-layer
in its entire depth, no impact on the hydrophobic lipid alkyl
lattice caused a decreased tendency to rupture in the presence
chains was observed [80-83]. Thus, S-layer lattices constitute
of �HL, indicating an enhanced stability [87]. Even single
unique supporting scaffoldings for lipid membranes [36, 56,
�HL pore recordings could be performed when reconstituted
in S-layer supported lipid membranes [88].
S-Layer Proteins as Key Components
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8 917
Main Phospholipid isolated from Thermoplasma acidophilum (7)
glycerol dialkyl nonitol tetraetherlipid extracted and purified
from Sulfolobus and Metallosphaera archaea (8)
OH OH OH OH OH OH
Fig. (8). Chemical structures of the phospholipid 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (6), the membrane-spanning tetraether-
lipids Main Phospholipid (7) isolated from Thermoplasma acidophilum, and glycerol dialkyl nonitol tetraetherlipid (8) extracted and purified
from Sulfolobus and Metallosphaera archaea.
The functionality of lipid membranes resting on S-layer
surrounded by sodium buffer with incorporated valinomycin,
covered filters and gold electrodes was demonstrated by the
a potassium-selective ion carrier, revealed a resistance in the
reconstitution of �HL and membrane-active peptides [89,
G�-range. In contrast, for the same membrane bathed in
90]. In a first study, gramicidin A was incorporated into
potassium buffer the resistance dropped almost three orders
tetraetherlipid monolayers, but also in phospholipid bilayers
of magnitude due to the valinomycin-mediated ion transport.
which were deposited on S-layer covered filters [89]. These
These results demonstrated that the biomimetic approach of
membranes revealed not only a remarkable stability, parti-
copying the supramolecular architecture of archaeal cell
cularly with an S-layer cover, but the most striking result
envelopes opened new possibilities for exploiting functional
was that high-resolution conductance measurements on
lipid membranes at meso- and macroscopic scale [92].
single gramicidin pores were feasible. In addition, for thevery first time, with filter supported lipid membranes, even
4. CONCLUSION AND FUTURE PERSPECTIVES
single pore recordings were performed on reconstituted
Basic and applied S-layer research has demonstrated that
nature provides most elegant examples for nanometer sized,
The functionality of lipid membranes resting on S-layer
molecular self-assembly systems. There are only a few
covered gold electrodes was demonstrated by the reconsti-
examples in nature where proteins reveal the intrinsic capa-
tution of alamethicin, gramicidin and valinomycin [90]. Due
bility to self-assemble into crystalline arrays, in suspension
to the formation of conductive alamethicin channels, the
and on a great variety of surfaces and interfaces. Since S-
membrane resistance dropped two orders of magnitude
layer lattices are highly anisotropic structures with signi-
whereas the capacitance was not altered. Partial inhibition of
ficant differences in the topography and physicochemical
the alamethicin channels with amiloride and analogues was
properties of the inner and outer surface, it was most
demonstrated, as increasing amounts of inhibitor gave rise to
important to copy nature's solution for assembling a secreted
an increased membrane resistance [90]. In addition, an SLM
protein on the cell surface into lattices with defined

Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
orientation. This biomimetic strategy is particularly essential
molecular construction kit (Fig. 9). Although, a broad
to ensure that crystallization of genetically engineered S-
spectrum of applications for S-layers has been developed, it
layer proteins occurred in defined orientation on solid
is expected that other areas will emerge particularly in areas
supports (metals, polymers, silicon wafers), lipid membranes,
where top-down and bottom-up strategies are commonly
liposomes, and a great variety of nanoparticles [62-64].
Another line for exploiting the unique features of S-
layers is directed to the use of lattices as support and
stabilizing structures for functionalized lipid films and
liposomes (Figs. 5 and 7). Again, composite, semifluid
SLMs are biomimetic structures copying the supramolecular
principle of archaeal cell envelopes or human or animal virus
envelopes optimized during biological evolution for a great
variety of functions [5, 19, 36, 92].
The numerous benefits generated by the attachment of
coherent S-layer lattices on lipid vesicles and mono- orbilayer membranes already triggered innovative approachesfor membrane biosensors, high through-put screening,diagnostics, and different lab-on-a-chip designs. S-liposomesrevealed high potentials for the development of new drug-
Fig. (9). Schematic drawing of an S-layer lattice (yellow
targeting, drug-delivery and transfection systems [11, 20, 38,
chessboard) with regular and well orientated functional molecules
40, 41, 43, 93].
(grey knights). The S-layer lattice of SbpA provides an area of up to
Moreover, S-layer self-assembly products have been
13 to 13 nm2 for each functional molecule.
demonstrated to be particularly well-suited for a geometri-cally defined covalent attachment of haptens and immuno-
genic or immuno-stimulating substances [94]. Most recently,
Financial support from the Austrian Science Fund (FWF,
a remarkable immuno-modulating capacity of S-layers was
projects 16295-B10 and 17170-B10), the Erwin-Schrödinger
demonstrated for a fusion protein comprising an S-layer
Society for Nanosciences, the FP6 EC STREP NASSAP
protein from a Bacillaceae and the major birch pollen
(project 13352), the Volkswagen Stiftung (project I/77710),
allergen Bet v 1 [67]. It is expected that innovative and
and the Air Force Office of Scientific Research, USA
highly specific immunogenic components with intrinsic
(AFOSR, project F49620-03-1-0222) is gratefully acknow-
targeting and delivery functionalities can be developed
combining recombinant S-layer proteins with the supra-
molecular construction principle of virus envelopes (Fig. 6).
Another attractiveness for S-layer self-assembly systems
Major birch pollen allergen
is seen for non-life science applications. Current state-of-the-
art methods for self-assembly of nanoparticle arrays thatgenerally involve bifunctional linkers, molecular recognition,
Variable domain of a heavy chain camel
or Langmuir-Blodgett techniques do not offer the control and
flexibility of the S-layer system. The S-layer approach for
cAb directed against the prostate-specific
the first time allows adjustable lattice constants and control
over template surface properties by chemical or geneticmodifications [5, 11, 15, 18] as required in molecular
electronics, biocatalysis, and non-linear optics.
Enhanced green fluorescent protein
Currently, there is a strong need to improve and develop
Green fluorescent protein
procedures for high resolution structural analysis ofmembrane proteins which can not be recrystallized to a
quality suitable for X-ray analysis studies. By using S-layer
fusion proteins, such target proteins could be forced into
Microspheres-based detoxification system
order arrays (Fig. 9) [61] accessible for structural analysis
involving established methods for image reconstruction such
as high resolution (cryo) electron microscopy, X-ray and
S-layer protein of Bacillus sphaericus
neutron reflectivity, and grazing incidence X-ray diffraction
[81]. Intrinsic in plane distortions of lattices formed by S-layer fusion proteins can be corrected following standard
S-layer protein of Geobacillus stearo-
procedures [23, 95].
S-layer protein o f Geobacillus stearo-
It is now evident that S-layers represent unique
thermophilus ATCC 12980
patterning elements or base plates for a complex supra-
S-Layer Proteins as Key Components
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8 919
Secondary cell wall polymer
Amos, L.A.; Henderson, R.; Unwin, P.N.T. Progr. Biophys. Mol.
Biol., 1982, 39, 183.
Scanning force microscopy
Györvary, E.S.; Stein, O.; Pum, D.; Sleytr, U.B. J. Microsc., 2003,
212, 300.
Crystalline bacterial cell surface layer
Karrasch, S.; Hegerl, R.; Hoh, J.;, Baumeister, W.; Engel, A. Proc.
Natl. Acad. Sci. USA, 1994, 91, 836.
Pum, D.; Sleytr, U.B. Supramol. Sci., 1995, 2, 193.
Müller, D.J.; Baumeister, W.; Engel, A. J. Bacteriol., 1996, 178,
S-layer coated liposome
Scheuring, S.; Stahlberg, H.; Chami, M.; Houssin, C.; Rigaud, J.L.;
S-layer supported lipid membrane
Engel, A. Mol. Microbiol., 2002, 44, 675.
Surface plasmon resonance
Stroh, C.M.; Ebner, A.; Geretschläger, M.; Freudenthaler, G.;Kienberger, F.; Kamruzzahan, A.S.M.; Smith-Gill, S.J.; Gruber,
Transmission electron microscopy
H.J.; Hinterdorfer, P. Biophys. J., 2004, 87, 1981.
Sleytr, U.B.; Messner, P. In Electron Microscopy of Subcellular
Variable domain of a heavy chain of a
Dynamics; Plattner, H. Ed.; CRC Press: Boca Raton, FL, 1989; pp
camel heavy chain antibody
Ilk, N.; Kosma, P.; Puchberger, M.; Egelseer, E.M.; Mayer, H.F.;
Synthetic analogue of the (IgG)-binding
Sleytr, U.B.; Sára, M. J. Bacteriol., 1999, 181, 7643.
Howorka, S.; Sára, M.; Wang, Y.; Kuen, B.; Sleytr, U.B.; Lubitz,
B-domain of protein A of Staphylococcus
W.; Bayley, H. J. Biol. Chem., 2000, 275, 37876.
Sára, M.; Dekitsch, C.; Mayer, H.F.; Egelseer, E.M.; Sleytr, U.B. J.
Bacteriol., 1998, 180, 4146.
Two copies of the Z-domain
Schuster, B.; Sleytr, U.B. Rev. Molec. Biotechnol., 2000, 74, 233.
Pum, D.; Weinhandl, M.; Hödl, C.; Sleytr, U.B. J. Bacteriol., 1993,
175, 2762.
Sleytr, U.B. Int. Rev. Cytol., 1978, 53, 1.
Küpcü, S.; Sára, M.; Sleytr, U.B. Biochim. Biophys. Acta, 1995,
Sleytr, U.B.; Messner, P.; Minnikin, D.E.; Heckels, J.E.; Virji M.;
1235, 263.
Russell, R.B.B. In Bacterial Cell Surface Techniques, Hancock, I.
Mader, C.; Küpcü, S.; Sára, M.; Sleytr, U.B. Biochim. Biophys.
C.; Poxton, I. Ed.; John Wileys & Son, Chichester, 1988; pp. 1-31.
Acta, 1999, 1418, 106.
Sleytr, U.B. FEMS Microbiol. Reviews 1997, 20, 5.
Mader, C.; Küpcü, S.; Sleytr, U.B.; Sára, M. Biochim. Biophys.
Sleytr, U.B.; Beveridge, T.J. Trends Microbiol., 1999, 7, 253.
Acta, 2000, 1463, 142.
Sleytr, U.B.; Messner, P.; Pum, D.; Sára, M. Angew. Chem. Int.
Toca-Herrera, J. L.; Krastev, R.; Bosio, V.; Küpcü, S.; Pum, D.;
Ed., 1999, 38, 1034.
Fery, A.; Sára, M.; Sleytr, U. B. Small, 2005, 1, 339.
König, H. Can. J. Microbiol., 1988, 34, 395.
Toca-Herrera, J. L.; Moreno-Flores, S.; Friedmann, J.; Pum, D.;
Beveridge, T.J. Int Rev Cytol., 1981, 72, 229.
Sleytr, U. B. Microsc. Res. Tech., 2004, 66, 163.
Baumeister, W.; Lembecke, G. J. Bioenerg. Biomembr., 1992, 24,
Sára, M.; Pum, D.; Schuster, B.; Sleytr, U.B. J. Nanosci.
Nanotechnol., 2005, 5, 1936.
Sára, M.; Sleytr, UB. J. Bacteriol., 2000, 182, 859.
Huber, C.; Ilk, N.; Rünzler, D.; Egelseer, E.-M.; Weigert, S.;
Messner, P.; Sleytr, U.B. Adv. Microb. Physiol., 1992, 33, 213.
Sleytr, U.B.; Sára, M. Mol. Microbiol., 2005, 55, 197.
Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B. In Supramolecular
Ilk, N.; Völlenkle, C.; Egelseer, E.-M.; Breitwieser, A.; Sleytr,
Polymers 2, Ciferri, A. Ed.; Marcel Dekker: New York, Basel,
U.B.; Sára, M. Appl. Environ. Microbiol., 2002, 68, 3251.
2005; pp. 583.
Jarosch, M.; Egelseer, E.-M.; Huber, C.; Moll, D.; Mattanovich, D.;
Sára, M.; Egenseer, E.-M. In Crystalline Bacterial Cell Surface
Sleytr, U.B.; Sára, M. Microbiology, 2001, 147, 1353.
Proteins, Sleytr, U.B.; Messner, P.; Pum, D.; Sára, M. Eds.;
Pavkov, T.; Oberer, M.; Egelseer, E.-M.; Sára, M.; Sleytr, U.B.;
Academic Press: Austin, 1996; pp. 103.
Keller, W. Acta Crystallogr. D Biol. Crystallogr., 2003, 59, 1466.
Sára, M. Trends Microbiol., 2001, 9, 47.
Steindl, C.; Schäffer, C.; Wugeditsch, T.; Graninger, M.; Matecko,
Sára, M.; Sleytr, U.B. J. Bacteriol., 1987, 169, 4092.
I.; Müller, N.; Messner, P. Biochem. J., 2002, 368, 483.
Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B.; Messner, P.;
Schäffer, C.; Messner, P. Microbioogy, 2005, 151, 643.
Schäffer, C. In Biopolymers, Steinbüchel, A.; Fahnestock, S. Eds.;
Cava, F.; de Pedro. M.A.; Schwarz, H.; Henne, A.; Berenguer, J.
Wiley-VCH: Weinheim, 2002; Vol. 7, pp. 285.
Mol. Microbiol., 2004, 52, 677.
Messner, P.; Pum, D.; Sára, M.; Stetter, K.O.; Sleytr, U.B. J.
Mesnage, S.; Fontaine, S.; Mignot, T.; Delepierre, M.; Mock, M.;
Bacteriol., 1986, 166, 1046.
Fouet, A. EMBO J., 2000, 19, 4473.
Pum, D.; Messner P.; Sleytr, U.B. J. Bacteriol., 1991, 173, 6865.
Egelseer, E.-M.; Leitner, K.; Jarosch, M.; Hotzy, C.; Zayni, S.;
Sleytr, U.B.; Pum, D.; Sára, M.; Schuster, B. In Encyclopedia of
Sleytr, U.B.; Sára, M. J. Bacteriol. 1998, 180, 1488.
Nanoscience and Nanotechnology, Nalwa, H.S. Ed.; Academic
Jarosch, M.; Egelseer, E.-M.; Mattanovich, D.; Sleytr, U.B.; Sára,
Press, San Diego, 2004; pp 693.
M. Microbiology, 2000, 146, 273.
Sleytr, U.B.; Egelseer, E.-M.; Pum, D.; Schuster, B. In:
Egelseer, E.-M.; Danhorn, T.; Pleschberger, M.; Hotzy, C.; Sleytr,
NanoBiotechnologie: Concepts, Methods and Perspectives,
U.B.; Sára, M. Arch. Microbiol., 2001, 177, 70.
Niemeyer, C. M.; Mirkin, C. A. Eds.; Wiley-VCH: Weinheim,
Messner, P.; Sleytr, U.B.; Christian, R.; Schulz, G.; Unger, F.M.
Germany, 2004, pp. 77.
Carbohydr. Res., 1987, 168, 211.
Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B. In Nano-surface
Schäffer, C.; Kählig, H.; Christian, R.; Schulz, G.; Zayni, S.;
chemistry, Rosoff, M. Ed.; Marcel Dekker, Inc.: New York, 2001,
Messner, P. Microbiology, 1999, 145, 1575.
Pum, D.; Schuster, B.; Sára, M.; Sleytr, U.B. IEE Proc.
Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B. Prog. Surf. Sci., 2001,
Nanobiotech., 2004, 151, 83.
68, 231.
Schuster, B.; Gufler, P.C.; Pum, D.; Sleytr, U.B. IEEE Trans.
Robards, A.W.; Sleytr, U.B. In Practical Methods in Electron
Nanobiosci., 2004, 3, 16.
Microscopy; Glauert, A.M. Ed.; Elsevier Sciences B. V: Amsterdam,
Bayley, H.; Braha, O.; Cheley, S.; Gu, L.-Q. In
1985; Vol. 10.
NanoBiotechnology: Concepts, Methods and Perspectives,
Baumeister, W.; Engelhardt, H. In Electron Microscopy of
Niemeyer, C.M.; Mirkin C.A. Eds.; Wiley-VCH Verlag:
Proteins; Harris J.R.; Horne, R.W. Eds.; Academic Press: London,
Weinheim; 2004; pp. 93-112.
1987; Vol. 6, pp. 109.
Huber, C.; Liu, L.; Egelseer, E.-M.; Moll, D.; Knoll, W.; Sleytr,
Hovmöller, S.; Sjögren, A.; Wang, D.N. Prog. Biophys. Mol. Biol.,
U.B.; Sára, M. Small, 2006, 2, 142.
1988, 51, 131.
Moll, D.; Huber, C.; Schlegel, B.; Pum, D.; Sleytr, U.B.; Sára, M.
Proc. Natl. Acad. Sci. USA, 2002, 99, 14646.
Mini-Reviews in Medicinal Chemistry, 2006, Vol. 6, No. 8
Schuster et al.
Pleschberger, M.; Neubauer, A.; Egelseer, E.-M.; Weigert, S.;
Pum, D.; Sleytr, U.B. Thin Solid Films, 1994, 244, 882.
Lindner, B.; Sleytr, U.B.; Muyldermans, S.; Sára, M. Bioconj.
Schuster, B.; Pum, D.; Sleytr, U.B. Biochim. Biophys. Acta, 1998,
Chem., 2003, 14, 440.
1369, 51.
Pleschberger, M.; Saerens, D.; Weigert, S.; Sleytr, U.B.;
Weygand, M.; Wetzer, B.; Pum, D.; Sleytr, U.B.; Cuvillier, N.;
Muyldermans, S.; Sára, M.; Egelseer, E.-M. Bioconj. Chem., 2004,
Kjaer, K.; Howes, P.B.; Lösche, M. Biophys. J., 1999, 76, 458.
15, 664.
Weygand, M.; Schalke, M.; Howes, P.B.; Kjaer, K.; Friedmann, J.;
Völlenkle, C.; Weigert, S.; Ilk, N.; Egelseer, E.-M.; Weber, V.;
Wetzer, B.; Pum, D.; Sleytr, U.B.; Lösche, M. J. Mater. Chem.,
Loth, F.; Falkenhagen, D.; Sleytr, U.B.; Sára, M. Appl. Environ.
2000, 10, 141.
Microbiol., 2004, 70, 1514. Highlighted in Nat. Rev. Microbiol.,
Weygand, M.; Kjaer, K.; Howes, P.B.; Wetzer, B.; Pum, D.; Sleytr,
2004, 2, 353.
U.B.; Lösche, M. J. Phys. Chem. B., 2002, 106, 5793
Weber, V.; Weigert, S.; Sára, M.; Sleytr, U.B.; Falkenhagen, D.
Schuster, B.; Gufler, P.C.; Pum, D.; Sleytr, U.B. Langmuir, 2003,
Ther. Apher., 2001, 5, 433.
19, 3393.
Breitwieser, A.; Egelseer, E.-M.; Ilk, N.; Moll, D.; Hotzy, C.;
Schuster, B.; Sleytr, U.B. Biochim. Biophys. Acta, 2002, 1563, 29.
Bohle, B.; Ebner, C.; Sleytr, U.B.; Sára, M. Protein Eng., 2002; 15,
Bhakdi, S.; Tranum-Jensen, J. Microbiol. Rev., 1991, 55, 733.
Schuster, B.; Pum, D.; Braha, O.; Bayley, H.; Sleytr, U.B. Biochim.
Bohle, B.; Breitwieser, A.; Zwölfer, B.; Jahn-Schmid, B.; Sára, M.;
Biophys. Acta, 1998, 1370, 280.
Sleytr, U.B.; Ebner, C. J. Immunol., 2004, 172, 6642.
Schuster, B.; Sleytr, U.B. Bioelectrochemistry, 2002, 55, 5.
Ilk, N.; Küpcü, S.; Moncayo, G.; Klimt, S.; Ecker, R.; Hofer-
Schuster, B.; Weigert, S.; Pum, D.; Sára, M.; Sleytr, U.B.
Warbinek, R.; Egelseer, E.-M.; Sleytr, U.B.; Sára, M. Biochem. J.,
Langmuir, 2003, 19, 2392.
2004, 370, 441.
Gufler, P.C.; Pum, D.; Sleytr, U.B.; Schuster, B. Biochim. Biophys.
Crommelin, D.; Storm, G. J. Liposome Res., 2003, 13, 33.
Acta, 2004, 1661, 154.
Lasic, D.D.; Papahadjopoulos, D. Science, 1995, 267, 1275.
Schuster, B.; Pum, D.; Sára, M.; Braha, O.; Bayley, H.; Sleytr, U.
Torchilin, V.P. Nat. Rev. Drug Discov., 2005, 4, 145.
B. Langmuir, 2001, 17, 499.
Lasic, D.D. Trends Biotechnol., 1998, 16, 307.
Schuster, B. NanoBiotechnology, 2005, 1, 153.
Templeton, N.S.; Lasic, D.D. Mol. Biotechnol., 1999, 11, 175.
Schuster, B.; Sleytr, U.B. In Advanves in Planar Lipid Bilayers and
Ostro, M.J.; Cullis, P.R. Am. J. Hosp. Pharm., 1989, 46, 1576.
Liposomes, Tien, T.H.; Ottova, A. Eds.; Elsevier Science:
Küpcü, S.; Lohner, K.; Mader, C.; Sleytr, U.B. Mol. Membr. Biol.,
Amsterdam, 2005; Vol. 1, pp. 247-293.
1998, 15, 69.
Jahn-Schmid, B.; Graninger, M.; Glozik, M.; Küpcü, S.; Ebner, C.;
Gerstein, M.; Hegyi, H. FEMS Microbiol. Rev., 1998, 22, 277.
Unger, F.M.; Sleytr, U.B.; Messner, P. Immunotechnology, 1996, 2,
Ellis, C.; Smith, A. Nature Rev. Drug Disc., 2004, 3, 237.
Wetzer, B.; Pfandler, A.; Györvary, E.; Pum, D.; Lösche, M.;
Crowther, R.A.; Sleytr, U.B. J. Ultrastruct. Res., 1977, 58, 41.
Sleytr, U.B. Langmuir, 1998, 14, 6899.
Received: November 10, 2005
Revised: January 17, 2006
Accepted: January 18, 2006
Source: http://homes.nano.aau.dk/fp/self-assembling/pdf-material/protein-drugdelivery.pdf
2015 Comprehensive Formulary HeartlandPlains Health 2015 Formulary List of Covered Drugs PLEASE READ: THIS DOCUMENT CONTAINS INFORMATION ABOUT THE DRUGS WE COVER IN THIS PLAN We have made no changes to this formulary since 10/15/15. For more recent information or other questions, please contact HeartlandPlains Health (HMO) Customer Service, at 1-866-792-0184or, for TTY users, 711, 8:00 am to 8:00 pm, Monday-Friday and 8:00 am to 8:00 pm, Monday-Sunday October 1 through February 14, or visit www.HeartlandPlainsHealth.com.
Seminario de Metodología de la Investigación Dr. José Manuel Carrillo Hernández UNIVERSIDAD AUTÓNOMA DE DURANGO DIVISIÓN DE ESTUDIOS DE POSGRADO CENTRO DE INVESTIGACIÓN Y DESARROLLO SEMINARIO DE METODOLOGÍA DE LA ELABORADO POR: DR. JOSÉ MANUEL CARRILLO HERNÁNDEZ INVESTIGADOR DEL CENTRO DE INVESTIGACIÓN Y DESARROLLO DE LA U.A.D.





